Active regulatory elements recruit cohesin to establish cell specific chromatin domains
- PMID: 40189615
- PMCID: PMC11973168
- DOI: 10.1038/s41598-025-96248-4
Active regulatory elements recruit cohesin to establish cell specific chromatin domains
Abstract
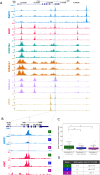
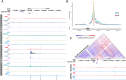
As the 3D structure of the genome is analysed at ever increasing resolution it is clear that there is considerable variation in the 3D chromatin architecture across different cell types. It has been proposed that this may, in part, be due to increased recruitment of cohesin to activated cis-elements (enhancers and promoters) leading to cell-type specific loop extrusion underlying the formation of new sub-TADs. Here we show that cohesin correlates well with the presence of active enhancers and that this varies in an allele-specific manner with the presence or absence of polymorphic enhancers which vary from one individual to another. Using the alpha globin cluster as a model, we show that when all enhancers are removed, peaks of cohesin disappear from these regions and the erythroid specific sub-TAD is no longer formed. Re-insertion of the major alpha globin enhancer (R2) is associated with re-establishment of recruitment and increased interactions. In complementary experiments insertion of the R2 enhancer element into a "neutral" region of the genome recruits cohesin, induces transcription and creates a new large (75 kb) erythroid-specific domain. Together these findings support the proposal that active enhancers recruit cohesin, stimulate loop extrusion and promote the formation of cell specific sub-TADs.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: J.R.H. is founder, shareholder, and paid consultant of Nucleome Therapeutics. J.R.H hold patents for Capture-C (WO2017068379A1, EP3365464B1, US10934578B2). T.A.M. is a shareholder in and consultant for Dark Blue Therapeutics. This author declares no other financial or non-financial interests. The remaining authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

