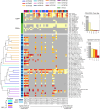
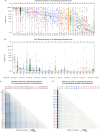
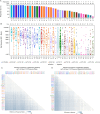
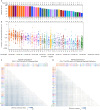
A polyvalent DNA prime with matched polyvalent protein/GLA-SE boost regimen elicited the most robust and broad IgG and IgG3 V1V2 binding antibody and CD4+ T cell responses among 13 HIV vaccine trials
- PMID: 40190112
- PMCID: PMC11980190
- DOI: 10.1080/22221751.2025.2485317
A polyvalent DNA prime with matched polyvalent protein/GLA-SE boost regimen elicited the most robust and broad IgG and IgG3 V1V2 binding antibody and CD4+ T cell responses among 13 HIV vaccine trials
Abstract
Developing an effective HIV vaccine is a momentous challenge. An exceptionally wide range of candidate HIV vaccines have been tested, yet many were poorly immunogenic, and of the select few that advanced into efficacy trials, only one demonstrated any efficacy. Here we report the results of the largest-scale cross-protocol immunogenicity comparison to date: 13 HIV vaccine trials (including 36 vaccine regimens) conducted across nine countries worldwide, strengthened by standardized trial designs, validated assays in centralized laboratories, and harmonized immunogenicity endpoints - providing an objective approach to identify the HIV vaccine candidate(s) with the best immunogenicity. A polyvalent DNA prime + protein boost regimen (HVTN 124) including Env immunogens of four subtypes, matched between prime and boost, achieved the best anti-V1V2 antibody responses by a large margin and also induced high CD4+ T-cell responses - two key immune responses implicated in HIV vaccine protection. Our results provide strong support to test this promising HIV vaccine design in more advanced phase clinical trials and will also guide the future design of additional HIV vaccines.Trial registration: ClinicalTrials.gov identifier: NCT01799954..Trial registration: ClinicalTrials.gov identifier: NCT02109354..Trial registration: ClinicalTrials.gov identifier: NCT02404311..Trial registration: ClinicalTrials.gov identifier: NCT02207920..Trial registration: ClinicalTrials.gov identifier: NCT02296541..Trial registration: ClinicalTrials.gov identifier: NCT03284710..Trial registration: ClinicalTrials.gov identifier: NCT02915016..Trial registration: ClinicalTrials.gov identifier: NCT02997969..Trial registration: ClinicalTrials.gov identifier: NCT03122223..Trial registration: ClinicalTrials.gov identifier: NCT03409276..Trial registration: ClinicalTrials.gov identifier: NCT02968849..Trial registration: ClinicalTrials.gov identifier: NCT03060629..Trial registration: ClinicalTrials.gov identifier: NCT00223080..
Keywords: Binding antibody multiplex assay; Env V1V2 binding antibody response breadth score; cross-protocol analysis; intracellular cytokine staining; matched polyvalent DNA-polyvalent protein/GLA-SE prime-boost regimen.
Conflict of interest statement
LNC was compensated for work on this manuscript through a consulting agreement with Fred Hutchinson Cancer Center. FT was an employee of Janssen/Johnson & Johnson at the time the work was conducted and owns stock in Johnson & Johnson. KM is a Member of the PrEPVACC TSC and a Member of the IDDO DAC (Infectious Disease data Observatory, Data Access Committee). Both are unpaid positions. L-GB has received honoraria in the past for advisories to Janssen/Johnson & Johnson, ViiV Healthcare, Gilead Sciences, and Merck PTY Ltd. None exceeded $2000 and all were unrelated to the topic of this manuscript. SL is a shareholder of Worcester HIV Vaccine.
Figures
References
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
