HIF-α signaling regulates the macrophage inflammatory response during Leishmania major infection
- PMID: 40191198
- PMCID: PMC11969800
- DOI: 10.3389/fimmu.2025.1487311
HIF-α signaling regulates the macrophage inflammatory response during Leishmania major infection
Abstract
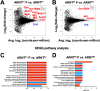
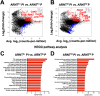
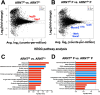
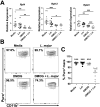
Cutaneous leishmaniasis (CL) contributes significantly to the global burden of neglected tropical diseases, with 12 million people currently infected with Leishmania parasites. CL encompasses a range of disease manifestations, from self-healing skin lesions to permanent disfigurations. Currently there is no vaccine available, and many patients are refractory to treatment, emphasizing the need for new therapeutic targets. Previous work demonstrated macrophage HIF-α-mediated lymphangiogenesis is necessary to achieve efficient wound resolution during murine L. major infection. Here, we investigate the role of macrophage HIF-α signaling independent of lymphangiogenesis. We sought to determine the relative contributions of the parasite and the host-mediated inflammation in the lesional microenvironment to myeloid HIF-α signaling. Because HIF-α activation can be detected in infected and bystander macrophages in leishmanial lesions, we hypothesize it is the host's inflammatory response and microenvironment, rather than the parasite, that triggers HIF-α activation. To address this, macrophages from mice with intact HIF-α signaling (LysMCreARNTf/+) or mice with deleted HIF-α signaling (LysMCreARNTf/f) were subjected to RNASequencing after L. major infection and under pro-inflammatory stimulus. We report that L. major infection alone is enough to induce some minor HIF-α-dependent transcriptomic changes, while infection with L. major in combination with pro-inflammatory stimuli induces numerous transcriptomic changes that are both dependent and independent of HIF-α signaling. Additionally, by coupling transcriptomic analysis with several pathway analyses, we found HIF-α suppresses pathways involved in protein translation during L. major infection in a pro-inflammatory environment. Together these findings show L. major induces a HIF-α-dependent transcriptomic program, but HIF-α only suppresses protein translation in a pro-inflammatory environment. Thus, this work indicates the host inflammatory response, rather than the parasite, largely contributes to myeloid HIF-α signaling during Leishmania infection.
Keywords: HIF - 1α; leishmania; leishmaniasis; macrophages; translation.
Copyright © 2025 Fry, Washam, Roys, Bowlin, Venugopal, Bird, Byrum and Weinkopff.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Update of
-
HIF-α signaling regulates the macrophage inflammatory response during Leishmania major infection.bioRxiv [Preprint]. 2024 Aug 27:2024.08.27.605844. doi: 10.1101/2024.08.27.605844. bioRxiv. 2024. Update in: Front Immunol. 2025 Mar 21;16:1487311. doi: 10.3389/fimmu.2025.1487311. PMID: 39253467 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

