Cardiomyocyte-Enriched USP20 Ameliorates Pathological Cardiac Hypertrophy by Targeting STAT3 Deubiquitination
- PMID: 40192103
- PMCID: PMC12199517
- DOI: 10.1002/advs.202416478
Cardiomyocyte-Enriched USP20 Ameliorates Pathological Cardiac Hypertrophy by Targeting STAT3 Deubiquitination
Abstract
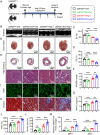
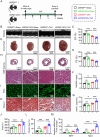
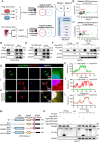
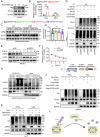
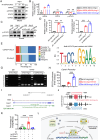
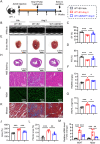
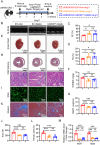
Although pathological cardiac hypertrophy is a key driver of heart failure, the underlying mechanisms remain incompletely elucidated. This study investigates the role and mechanism of deubiquitinating enzyme (DUB) ubiquitin-specific protease 20 (USP20) in cardiac hypertrophy. Transcriptomic profiling of hypertrophic hearts shows significant alterations in the expression of DUBs, including a remarkable downregulation of USP20. USP20 is predominantly expressed in cardiomyocytes. Co-immunoprecipitation (Co-IP) followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) is used to identify USP20 substrates. Cleavage Under Targets and Tagmentation assay (CUT&Tag) sequencing is employed to identify downstream targets of signal transducer and activator of transcription 3 (STAT3). Functionally, USP20 deficiency exacerbates cardiac hypertrophy induced by either angiotensin II (Ang II) or transverse aortic constriction (TAC), whereas USP20 overexpression alleviates hypertrophic responses. Mechanistically, USP20 deubiquitinates STAT3 by removing K63-linked ubiquitin chains at K177 via its H645 active site, reducing STAT3 phosphorylation and nuclear translocation. This inhibites STAT3's transcriptional activity at coactivator-associated arginine methyltransfer (Carm1) promoter, leading to upregulated CARM1 expression and mitigated hypertrophy. Importantly, the STAT3 inhibitor Stattic confirms STAT3 serves as a key substrate mediating the cardiac protective effects of USP20. These findings unveil a novel USP20/STAT3/CARM1 axis in cardiomyocytes and reveal its therapeutic potential for cardiac hypertrophy.
Keywords: cardiac hypertrophy; cardiomyocyte; deubiquitinating enzyme; signal transducer and activator of transcription 3; ubiquitin‐specific protease 20.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Shimizu I., Minamino T., J. Mol. Cell. Cardiol. 2016, 97, 245. - PubMed
-
- Metra M., Teerlink J. R., Lancet 2017, 390, 1981. - PubMed
-
- Maron B. J., Rowin E. J., Maron M. S., Annu. Rev. Med. 2022, 73, 363. - PubMed
-
- Tham Y. K., Bernardo B. C., Ooi J. Y., Weeks K. L., McMullen J. R., Arch. Toxicol. 2015, 89, 1401. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
