Subtyping Burkitt Lymphoma by DNA Methylation
- PMID: 40192513
- PMCID: PMC11974478
- DOI: 10.1002/gcc.70042
Subtyping Burkitt Lymphoma by DNA Methylation
Abstract
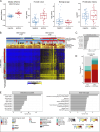
Burkitt lymphoma (BL) is an aggressive germinal center B-cell-derived malignancy. Historically, sporadic, endemic, and immunodeficiency-associated variants were distinguished, which differ in the frequency of Epstein-Barr virus (EBV) positivity. Aiming to identify subgroups based on DNA methylation patterns, we here profiled 96 BL cases, 17 BL cell lines, and six EBV-transformed lymphoblastoid cell lines using Illumina BeadChip arrays. DNA methylation analyses clustered the cases into four subgroups: two containing mostly EBV-positive cases (BL-mC1, BL-mC2) and two containing mostly EBV-negative cases (BL-mC3, BL-mC4). The subgroups BL-mC1/2, enriched for EBV-positive cases, showed increased DNA methylation, epigenetic age, and, in part, proliferation history compared to BL-mC3/4. CpGs hypermethylated in EBV-positive BLs were enriched for polycomb repressive complex 2 marks, while the CpGs hypomethylated in EBV-negative BLs were linked to, for example, B-cell receptor signaling. EBV-associated hypermethylation affected regulatory regions of genes frequently mutated in BL (e.g., CCND3, TP53) and impacted superenhancers. This finding suggests that hypermethylation may compensate for the lower mutational burden of pathogenic drivers in EBV-positive BLs. Though minor, significant differences were also observed between EBV-positive endemic and sporadic cases (e.g., at the SOX11 and RUNX1 loci). Our findings suggest that EBV status, rather than epidemiological variants, drives the DNA methylation-based subgrouping of BL.
Keywords: Africa; Burkitt lymphoma; DNA methylation; Epstein–Barr virus; immunodeficiency.
© 2025 The Author(s). Genes, Chromosomes and Cancer published by Wiley Periodicals LLC. This article has been contributed to by U.S. Government employees and their work is in the public domain in the USA.
Conflict of interest statement
M.J.B. is currently an employee of Swedish Orphan Biovitrum A.B. The other authors declare no conflicts of interest.
Figures


References
-
- Aukema S. M., Theil L., Rohde M., et al., “Sequential Karyotyping in Burkitt Lymphoma Reveals a Linear Clonal Evolution With Increase in Karyotype Complexity and a High Frequency of Recurrent Secondary Aberrations,” British Journal of Haematology 170, no. 6 (2015): 814–825, 10.1111/bjh.13501. - DOI - PubMed
MeSH terms
Grants and funding
- PID20210125017OB-I00/Spanish Ministry of Science, Innovation and University
- MICIU/AEI/10.13039/501100011033/Spanish Ministry of Science, Innovation and University
- 01KU1002A-J/German Ministry of Science and Education
- 036166B/German Ministry of Science and Education
- 01KU1505E/German Ministry of Science and Education
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

