Single-Cell Transcriptomic Landscape Deciphers Intratumoral Heterogeneity and Subtypes of Acral and Mucosal Melanomas
- PMID: 40192737
- PMCID: PMC12163602
- DOI: 10.1158/1078-0432.CCR-24-3164
Single-Cell Transcriptomic Landscape Deciphers Intratumoral Heterogeneity and Subtypes of Acral and Mucosal Melanomas
Abstract
Purpose: To identify the specific intratumoral and microenvironmental heterogeneity of acral melanoma (AM) and mucosal melanoma (MM), we aimed to delineate their distinct cellular compositions, evolutionary trajectories, and subtype-specific therapeutic strategies.
Experimental design: Single-cell transcriptomic and genomic landscapes were analyzed across 42 melanoma (28 AM, 11 MM, and 3 nonacral cutaneous melanoma) samples, supplemented by in vitro and in vivo validation. Tumor and stromal cells were profiled using single-cell RNA sequencing, whole-exome sequencing, and functional assays, including transwell migration, co-culture systems, and xenograft models.
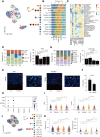
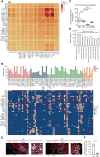
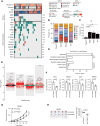
Results: Tumor cells exhibited divergent evolutionary routes, with MM dominated by MGP+/PCOLCE+ subpopulations showing high epithelial-to-mesenchymal transition potential. MM displayed elevated neutrophil infiltration and CXCL3+ tumor-associated macrophages, whereas AM was enriched with PI16+ cancer-associated fibroblasts promoting tumor proliferation. Molecular classification revealed MM subtypes: an antigen-presenting subtype linked to favorable outcomes and a proliferative subtype associated with recurrence. TIGIT+ regulatory T cells were enriched in AM, suggesting targeted inhibition potential. Genomic analysis connected BRAF/NRAS mutations to ALDOA+ stem-like tumor cells and identified prostaglandin D2 synthetase as a therapeutic target in triple-wild-type/melanomas.
Conclusions: Our study provides a comprehensive comparison of AM and MM, uncovering subtype-specific stromal-immune interactions and molecular programs. The findings highlight actionable targets (e.g., TIGIT in AM and CXCL3+ macrophages in MM) and propose a framework for precision therapies, biomarker-driven trials, and risk stratification to improve outcomes in these aggressive melanomas.
©2025 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
No disclosures were reported.
Figures




References
-
- de Visser KE, Joyce JA. The evolving tumor microenvironment: from cancer initiation to metastatic outgrowth. Cancer Cell 2023;41:374–403. - PubMed
-
- Kalaora S, Nagler A, Wargo JA, Samuels Y. Mechanisms of immune activation and regulation: lessons from melanoma. Nat Rev Cancer 2022;22:195–207. - PubMed
-
- Bald T, Quast T, Landsberg J, Rogava M, Glodde N, Lopez-Ramos D, et al. Ultraviolet-radiation-induced inflammation promotes angiotropism and metastasis in melanoma. Nature 2014;507:109–13. - PubMed
-
- Lian B, Cui CL, Zhou L, Song X, Zhang XS, Wu D, et al. The natural history and patterns of metastases from mucosal melanoma: an analysis of 706 prospectively-followed patients. Ann Oncol 2017;28:868–73. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

