Topology-driven negative sampling enhances generalizability in protein-protein interaction prediction
- PMID: 40193392
- PMCID: PMC12080959
- DOI: 10.1093/bioinformatics/btaf148
Topology-driven negative sampling enhances generalizability in protein-protein interaction prediction
Abstract
Motivation: Unraveling the human interactome to uncover disease-specific patterns and discover drug targets hinges on accurate protein-protein interaction (PPI) predictions. However, challenges persist in machine learning (ML) models due to a scarcity of quality hard negative samples, shortcut learning, and limited generalizability to novel proteins.
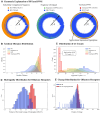
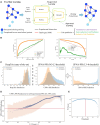
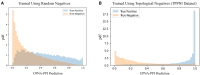
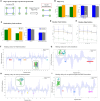
Results: In this study, we introduce a novel approach for strategic sampling of protein-protein noninteractions (PPNIs) by leveraging higher-order network characteristics that capture the inherent complementarity-driven mechanisms of PPIs. Next, we introduce Unsupervised Pre-training of Node Attributes tuned for PPI (UPNA-PPI), a high throughput sequence-to-function ML pipeline, integrating unsupervised pre-training in protein representation learning with Topological PPNI (TPPNI) samples, capable of efficiently screening billions of interactions. By using our TPPNI in training the UPNA-PPI model, we improve PPI prediction generalizability and interpretability, particularly in identifying potential binding sites locations on amino acid sequences, strengthening the prioritization of screening assays and facilitating the transferability of ML predictions across protein families and homodimers. UPNA-PPI establishes the foundation for a fundamental negative sampling methodology in graph machine learning by integrating insights from network topology.
Availability and implementation: Code and UPNA-PPI predictions are freely available at https://github.com/alxndgb/UPNA-PPI.
© The Author(s) 2025. Published by Oxford University Press.
Figures


References
-
- Abboud A, Khoury S, Leibowitz O et al. Listing 4-cycles. https://arxiv.org/abs/2211.10022, 2022, preprint: not peer reviewed.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

