Test allocation based on risk of infection from first and second order contact tracing
- PMID: 40193399
- PMCID: PMC11975095
- DOI: 10.1371/journal.pone.0320291
Test allocation based on risk of infection from first and second order contact tracing
Abstract
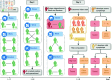
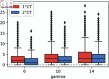
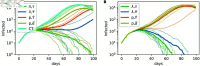
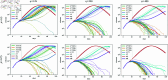
Strategies such as testing, contact tracing, and quarantine have been proven to be essential mechanisms to mitigate the propagation of infectious diseases. However, when an epidemic spreads rapidly and/or the resources to contain it are limited (e.g., not enough tests available on a daily basis), to test and quarantine all the contacts of detected individuals is impracticable. In this direction, we propose a method to compute the individual risk of infection over time, based on the partial observation of the epidemic spreading through the population contact network. We define the risk of individuals as their probability of getting infected from any of the possible chains of transmission up to length-two, originating from recently detected individuals. Ranking individuals according to their risk of infection can serve as a decision-making tool to prioritise testing, quarantine, or other preventive measures. We evaluate interventions based on our risk ranking through simulations using a fairly realistic agent-based model calibrated for COVID-19 epidemic outbreak. We consider different scenarios to study the role of key quantities such as the number of daily available tests, the contact tracing time-window, the transmission probability per contact (constant versus depending on multiple factors), and the age since infection (for varying infectiousness). We find that, when there is a limited number of daily tests available, our method is capable of mitigating the propagation more efficiently than some other approaches in the recent literature on the subject. A crucial aspect of our method is that we provide an explicit formula for the risk, avoiding the large number of iterations required to achieve convergence for the algorithms proposed in the literature. Furthermore, neither the entire contact network nor a centralised setup is required. These characteristics are essential for the practical implementation using contact tracing applications.
Copyright: © 2025 Soler et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



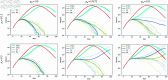
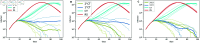
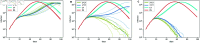
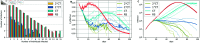
References
-
- Herbrich R, Rastogi R, Vollgraf R. CRISP: a probabilistic model for individual-level COVID-19 infection risk estimation based on contact data. arXiv. 2022. https://arxiv.org/abs/2006.04942
-
- Batlle P, Bruna J, Fernandez-Granda C, Preciado VM. Adaptive test allocation for outbreak detection and tracking in social contact networks. SIAM J Control Optim. 2022;60(2):S274–93. doi: 10.1137/20m1377874 - DOI
-
- Romijnders R, Asano YM, Louizos C, Welling M. No time to waste: practical statistical contact tracing with few low-bit messages. In Proceedings of The 26th International Conference on Artificial Intelligence and Statistics, PMLR; 2023. pp. 7943–60. Available: https://proceedings.mlr.press/v206/romijnders23a.html.
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

