Two redox-responsive LysR-type transcription factors control the oxidative stress response of Agrobacterium tumefaciens
- PMID: 40193708
- PMCID: PMC11975290
- DOI: 10.1093/nar/gkaf267
Two redox-responsive LysR-type transcription factors control the oxidative stress response of Agrobacterium tumefaciens
Abstract
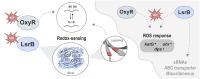
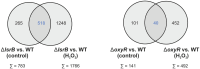
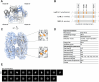
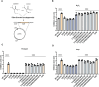
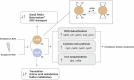
Pathogenic bacteria often encounter fluctuating reactive oxygen species (ROS) levels, particularly during host infection, necessitating robust redox-sensing mechanisms for survival. The LysR-type transcriptional regulator (LTTR) OxyR is a widely conserved bacterial thiol-based redox sensor. However, members of the Rhizobiales also encode LsrB, a second LTTR with potential redox-sensing function. This study explores the roles of OxyR and LsrB in the plant-pathogen Agrobacterium tumefaciens. Through single and combined deletions, we observed increased H2O2 sensitivity, underscoring their function in oxidative defense. Genome-wide transcriptome profiling under H2O2 exposure revealed that OxyR and LsrB co-regulate key antioxidant genes, including katG, encoding a bifunctional catalase/peroxidase. Agrobacterium tumefaciens LsrB possesses four cysteine residues potentially involved in redox sensing. To elucidate the structural basis for redox-sensing, we applied single-particle cryo-EM (cryogenic electron microscopy) to experimentally confirm an AlphaFold model of LsrB, identifying two proximal cysteine pairs. In vitro thiol-trapping coupled with mass spectrometry confirmed reversible thiol modifications of all four residues, suggesting a functional role in redox regulation. Collectively, these findings reveal that A. tumefaciens employs two cysteine-based redox sensing transcription factors, OxyR and LsrB, to withstand oxidative stress encountered in host and soil environments.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

