AgrC biotinylation inhibits Staphylococcus aureus infection
- PMID: 40193824
- PMCID: PMC11991674
- DOI: 10.1371/journal.pone.0318695
AgrC biotinylation inhibits Staphylococcus aureus infection
Abstract
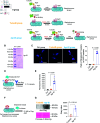
Staphylococcus aureus (S. aureus) is a leading cause of nosocomial infections, particularly among antibiotic-resistant strains. S. aureus virulence is governed by the accessory gene regulator (Agr) quorum sensing (QS) system, which relies on AgrC, a two-component histidine kinase, to detect secreted auto-inducing peptides (AIPs). Emerging evidence highlights the potential of inhibiting the interaction between AgrC and AIPs as a promising therapeutic strategy. Given the limited clinic methods in inhibiting AgrC, we hereby report a novel method utilizing TurboID, an engineered biotin ligase, to inhibit Agr C on S. aureus via its biotinylation. To achieve this goal, a fusion protein named TurboID-AgrD[Formula: see text] (Agr-ID) was designed to include an AgrC binding domain (AgrID[Formula: see text]) and a catalytic domain (TurboID) for AgrC biotinylation. By incubating with Alexa Fluor 647-conjugated streptavidin, the biotinylated AgrC on S. aureus was successfully visualized through fluorescence microscopy with 100x objective. We further confirmed the specific biotinylation of AgrC using Western Blotting, and biotinylated AgrC resulted in inhibiting the growth of S. aureus strains, including S. aureus 25923, S. aureus 43300, and S. aureus 6538 (MRSA). The downstream biological effect of AgrC biotinylation exhibited decreased virulence protein generation as monitored by the lower presence of apoptotic HEK 293T cells after incubating with S. aureus cell lysates and supernatant. The impaired colonizing features from biotinylated S. aureus 6538 were investigated by calculating the decreased ratio of cell death versus live HeLa cells. By further investigating the efficiency of the immune clearance of biotinylated S. aureus by mouse macrophages, we observed the enhanced uptake of S. aureus by murine macrophages in vivo. Overall, our work reveals that the biotinylation of AgrC can inhibit the growth and toxicity of S. aureus while simultaneously promoting the clearance of biotinylated S. aureus via macrophage phagocytosis.
Copyright: © 2025 Qian et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

