Exposure to Long- and Short-Chain Per- and Polyfluoroalkyl Substances in Mice and Ovarian-Related Outcomes: An in Vivo and in Vitro Study
- PMID: 40194260
- PMCID: PMC12120842
- DOI: 10.1289/EHP14876
Exposure to Long- and Short-Chain Per- and Polyfluoroalkyl Substances in Mice and Ovarian-Related Outcomes: An in Vivo and in Vitro Study
Abstract
Background: The extensive use of per- and polyfluoroalkyl substances (PFAS) has led to environmental contamination and bioaccumulation of these substances. Previous research linked PFAS exposure to female reproductive disorders, but the mechanism remains elusive. Further, most studies focused on legacy long-chain PFOA and PFOS, yet the reproductive impacts of other long-chain PFAS and short-chain alternatives are rarely explored.
Objectives: We investigated the effects of long- and short-chain PFAS on the mouse ovary and further evaluated the toxic mechanisms of long-chain perfluorononanoic acid (PFNA).
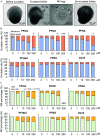
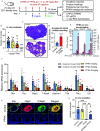
Methods: A 3D in vitro mouse ovarian follicle culture system and an in vivo mouse model were used, together with approaches of reverse transcription-quantitative polymerase chain reaction (RT-qPCR), enzyme-linked immunosorbent assay (ELISA), RNA sequencing (RNA-seq), pharmacological treatments, in situ zymography, histology, in situ hybridization, analytical chemistry, and benchmark dose modeling (BMD). Using these approaches, a wide range of exposure levels () of long-chain PFAS (PFOA, PFOS, PFNA) and short-chain PFAS (PFHpA, PFBS, GenX) were first tested in cultured follicles to examine their effects on follicle growth, hormone secretion, and ovulation. We identified as the most effective concentration for further investigation into the toxic mechanisms of PFNA, followed by an in vivo mouse exposure model to verify the accumulation of PFNA in the ovary and its ovarian-disrupting effects.
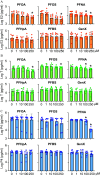
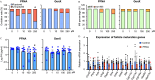
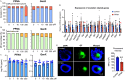
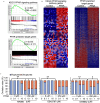
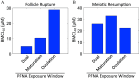
Results: In vitro cultured ovarian follicles exposed to long- but not short-chain PFAS showed poorer gonadotropin-dependent follicle growth, ovulation, and hormone secretion in comparison with control follicles. RT-qPCR and RNA-seq analyses revealed significant alterations in the expression of genes involved in follicle-stimulating hormone (FSH)-dependent follicle growth, luteinizing hormone (LH)-stimulated ovulation, and associated regulatory pathways in the PFNA-exposed group in comparison with the control group. The PPAR agonist experiment demonstrated that a peroxisome proliferator-activated receptor gamma () antagonist could reverse both the phenotypic and genotypic effects of PFNA exposure, restoring them to levels comparable to the control group. Furthermore, in vivo experiments confirmed that PFNA could accumulate in ovarian tissues and validated the in vitro findings. The BMD, in vitro, and in vivo extrapolation analyses estimated follicular rupture as the most sensitive end point and that observed effects occurred in the range of human exposure to long-chain PFAS.
Discussion: Our study demonstrates that long-chain PFAS, particularly PFNA, act as a agonist in granulosa cells to interfere with gonadotropin-dependent follicle growth, hormone secretion, and ovulation; and exposure to high levels of PFAS may cause adverse ovarian outcomes. https://doi.org/10.1289/EHP14876.
Figures








References
-
- Gebbink WA, van Asseldonk L, van Leeuwen SPJ. 2017. Presence of emerging per- and polyfluoroalkyl substances (PFASs) in river and drinking water near a fluorochemical production plant in the Netherlands. Environ Sci Technol 51(19):11057–11065, PMID: 28853567, 10.1021/acs.est.7b02488. - DOI - PMC - PubMed
-
- Gangal SV. 2004. Perfluorinated polymers. In: Kirk-Othmer Encyclopedia of Chemical Technology. Hoboken, NJ: John Wiley & Sons, Inc., 1–68.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
