CoPPIs algorithm: a tool to unravel protein cooperative strategies in pathophysiological conditions
- PMID: 40194557
- PMCID: PMC11975363
- DOI: 10.1093/bib/bbaf146
CoPPIs algorithm: a tool to unravel protein cooperative strategies in pathophysiological conditions
Abstract
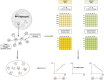
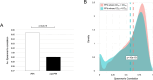
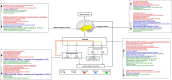
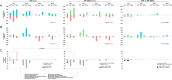
We present here the co-expressed protein-protein interactions algorithm. In addition to minimizing correlation-causality imbalance and contextualizing protein-protein interactions to the investigated systems, it combines protein-protein interactions and protein co-expression networks to identify differentially correlated functional modules. To test the algorithm, we processed a set of proteomic profiles from different brain regions of controls and subjects affected by idiopathic Parkinson's disease or carrying a GBA1 mutation. Its robustness was supported by the extraction of functional modules, related to translation and mitochondria, whose involvement in Parkinson's disease pathogenesis is well documented. Furthermore, the selection of hubs and bottlenecks from the weightedprotein-protein interactions networks provided molecular clues consistent with the Parkinson pathophysiology. Of note, like quantification, the algorithm revealed less variations when comparing disease groups than when comparing diseased and controls. However, correlation and quantification results showed low overlap, suggesting the complementarity of these measures. An observation that opens the way to a new investigation strategy that takes into account not only protein expression, but also the level of coordination among proteins that cooperate to perform a given function.
Keywords: PPI network; Parkinson; co-expression network; proteomics; topology.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
All authors declare that they have no conflicts of interest.
Figures



Similar articles
-
Construction and analysis of the protein-protein interaction networks based on gene expression profiles of Parkinson's disease.PLoS One. 2014 Aug 29;9(8):e103047. doi: 10.1371/journal.pone.0103047. eCollection 2014. PLoS One. 2014. PMID: 25170921 Free PMC article.
-
GBA1-Associated Parkinson's Disease Is a Distinct Entity.Int J Mol Sci. 2024 Jun 28;25(13):7102. doi: 10.3390/ijms25137102. Int J Mol Sci. 2024. PMID: 39000225 Free PMC article. Review.
-
Disturbed Mitochondria-Lysosome Crosstalk in GBA1-Associated Parkinson's Disease.Mov Disord. 2021 Aug;36(8):1792. doi: 10.1002/mds.28693. Epub 2021 Jul 1. Mov Disord. 2021. PMID: 34196414 No abstract available.
-
Reduced glucocerebrosidase is associated with increased α-synuclein in sporadic Parkinson's disease.Brain. 2014 Mar;137(Pt 3):834-48. doi: 10.1093/brain/awt367. Epub 2014 Jan 28. Brain. 2014. PMID: 24477431 Free PMC article.
-
The biochemical basis of interactions between Glucocerebrosidase and alpha-synuclein in GBA1 mutation carriers.J Neurochem. 2020 Jul;154(1):11-24. doi: 10.1111/jnc.14968. Epub 2020 Feb 3. J Neurochem. 2020. PMID: 31965564 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

