Multiple molecular diagnoses identified through genome sequencing in individuals with suspected rare disease
- PMID: 40195116
- PMCID: PMC12033986
- DOI: 10.1016/j.xhgg.2025.100430
Multiple molecular diagnoses identified through genome sequencing in individuals with suspected rare disease
Abstract
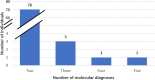
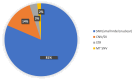
Genome sequencing is a powerful and comprehensive test that detects multiple variants of different types. The interrogation of variants across the genome enables the identification of multiple molecular diagnoses (MMDs) in a single individual. In this retrospective study, we describe individuals in whom MMDs were associated with the proband's indication for testing (IFT), secondary findings, or incidental findings. An MMD is considered where at least one of the findings is associated with the primary IFT and all variants are classified as either likely pathogenic or pathogenic. Clinical genome sequencing was performed for all individuals as part of the iHope program at the Illumina Laboratory Services between September 2017 and December 2023. The iHope cohort included 1,846 affected individuals, with 872 (47.2%) found to have at least one likely pathogenic or pathogenic variant associated with the primary IFT. Of these, 81 (9.3%) individuals had multiple clinically significant molecular findings, including 76 individuals with reported variants associated with 2 different conditions, and 5 individuals with more than 2 molecular findings. A total of 32 individuals (3.7%) had at least 2 molecular diagnoses related to the primary IFT, while in 49 (5.6%) individuals, the variant(s) reported for the second condition constituted a secondary or incidental finding. Our study highlights that among individuals with a likely pathogenic or pathogenic finding identified through genome sequencing, 9% have MMDs, which may have been missed with different testing methods. Of note, approximately 60% of the 81 individuals with an MMD had a potentially actionable secondary or incidental finding.
Keywords: WGS; incidental finding; multiple molecular diagnoses; rare disease; secondary finding.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.J.C., A.K., A.M., D.L.P., E.T., R.J.T., R.R., and the Illumina Laboratory Services Interpretation and Reporting Team are either current or former employees and shareholders at Illumina, Inc.
Figures

References
-
- Thorpe E., Williams T., Shaw C., Chekalin E., Ortega J., Robinson K., Button J., Jones M.C., Campo M.D., Basel D., et al. The impact of clinical genome sequencing in a global population with suspected rare genetic disease. Am. J. Hum. Genet. 2024;111:1271–1281. doi: 10.1016/j.ajhg.2024.05.006. - DOI - PMC - PubMed
-
- Stranneheim H., Lagerstedt-Robinson K., Magnusson M., Kvarnung M., Nilsson D., Lesko N., Engvall M., Anderlid B.-M., Arnell H., Johansson C.B., et al. Integration of whole genome sequencing into a healthcare setting: high diagnostic rates across multiple clinical entities in 3219 rare disease patients. Genome Med. 2021;13:40. doi: 10.1186/s13073-021-00855-5. - DOI - PMC - PubMed
-
- Ewans L.J., Minoche A.E., Schofield D., Shrestha R., Puttick C., Zhu Y., Drew A., Gayevskiy V., Elakis G., Walsh C., et al. Whole exome and genome sequencing in mendelian disorders: a diagnostic and health economic analysis. Eur. J. Hum. Genet. 2022;30:1121–1131. doi: 10.1038/s41431-022-01162-2. - DOI - PMC - PubMed
-
- Nurchis M.C., Altamura G., Riccardi M.T., Radio F.C., Chillemi G., Bertini E.S., Garlasco J., Tartaglia M., Dallapiccola B., Damiani G. Whole genome sequencing diagnostic yield for paediatric patients with suspected genetic disorders: systematic review, meta-analysis, and GRADE assessment. Arch. Public Health. 2023;81:93. doi: 10.1186/s13690-023-01112-4. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

