Enabling new insights from old scans by repurposing clinical MRI archives for multiple sclerosis research
- PMID: 40195318
- PMCID: PMC11976987
- DOI: 10.1038/s41467-025-58274-8
Enabling new insights from old scans by repurposing clinical MRI archives for multiple sclerosis research
Abstract
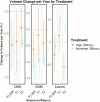
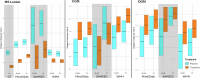
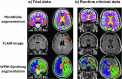
Magnetic resonance imaging (MRI) biomarkers are vital for multiple sclerosis (MS) clinical research and trials but quantifying them requires multi-contrast protocols and limits the use of abundant single-contrast hospital archives. We developed MindGlide, a deep learning model to extract brain region and white matter lesion volumes from any single MRI contrast. We trained MindGlide on 4247 brain MRI scans from 2934 MS patients across 592 scanners, and externally validated it using 14,952 scans from 1,001 patients in two clinical trials (primary-progressive MS and secondary-progressive MS trials) and a routine-care MS dataset. The model outperformed two state-of-the-art models when tested against expert-labelled lesion volumes. In clinical trials, MindGlide detected treatment effects on T2-lesion accrual and cortical and deep grey matter volume loss. In routine-care data, T2-lesion volume increased with moderate-efficacy treatment but remained stable with high-efficacy treatment. MindGlide uniquely enables quantitative analysis of archival single-contrast MRIs, unlocking insights from untapped hospital datasets.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: D.C. is a consultant for Hoffmann-La Roche. In the last three years he has been a consultant for Biogen, has received research funding from Hoffmann-La Roche, the International Progressive MS Alliance, the MS Society, the Medical Research Council, and the National Institute for Health Research (NIHR) University College London Hospitals (UCLH) Biomedical Research Centre, and a speaker’s honorarium from Novartis. He co-supervises a clinical fellowship at the National Hospital for Neurology and Neurosurgery, London, which is supported by Merck. F.B. acts as a member of the steering committee or Data Safety Monitoring Board for Biogen, Merck, ATRI/ACTC and Prothena. Consultant for Roche, Celltrion, Rewind Therapeutics, Merck, IXICO, Jansen, Combinostics. Research agreements with Merck, Biogen, GE Healthcare, Roche. Co-founder and shareholder of Queen Square Analytics LTD. O.C. is a NIHR Research Professor (RP-2017-08-ST2-004); over the last 2 years, member of independent DSMB for Novartis; she gave a teaching talk in a Merck local symposium, and contributed to an Advisory Board for Biogen; she is Deputy Editor of Neurology, for which she receives an honorarium; she has received research grant support from the MS Society of Great Britain and Northern Ireland, the NIHR UCLH Biomedical Research Centre, the Rosetree Trust, the National MS Society, and the NIHR-HTA. C.H. reports grant support from the MRC and MS Society. She has served as a consultant to Novartis, Roche, UCB and Sanofi. S.N. has received research funding from the Canadian Institutes of Health Research, the International Progressive MS Alliance, the Myelin Repair Foundation, Immunotec, and F. Hoffman LaRoche, not related to the current work; he is a consultant for Sana Biotech, has received a speaker’s honorarium from Novartis Canada, and is a part-time employee of NeuroRx Research. In the last 3 years, J.C. has received support from the Health Technology Assessment (HTA) Programme (National Institute for Health Research, NIHR), the UK MS Society, the US National MS Society and the Rosetrees Trust. He is supported in part by the NIHR University College London Hospitals (UCLH) Biomedical Research Centre, London, UK. He has been a local principal investigator for a trial in MS funded by MS Canada. A local principal investigator for commercial trials funded by: Ionis and Roche; and has taken part in advisory boards/consultancy for Biogen, Contineum Therapeutics, InnoCare, Lucid, Merck, NervGen, Novartis and Roche. G.J.M.P. is a shareholder and director of, and receives salary from, Bioxydyn Limited. He is a shareholder and director of Queen Square Analytics Limited. He is a shareholder and director of Quantitative Imaging Limited. D.C.A. is a shareholder and director of Queen Square Analytics Limited. In the past three years, A.E. has received research grants from the Medical Research Council (MRC), NHS England, Imperial College Healthcare Trust, National Institute for Health and Social Care Research (NIHR), Innovate UK, Biogen, Merck, and Roche. He has served as an advisory board member of Merck Serono and Bristol Myers Squib. He is the founder and equity stakeholder in Queen Square Analytics Limited. He serves on the editorial board of Neurology (American Academy of Neurology). The remaining authors declare no competing interests.
Figures





References
-
- Cardoso, M. J. et al. MONAI: An open-source framework for deep learning in healthcare. 10.48550/ARXIV.2211.02701 (2022)
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

