CRISPR-Cas9 genetic screens reveal regulation of TMPRSS2 by the Elongin BC-VHL complex
- PMID: 40195420
- PMCID: PMC11976923
- DOI: 10.1038/s41598-025-95644-0
CRISPR-Cas9 genetic screens reveal regulation of TMPRSS2 by the Elongin BC-VHL complex
Abstract
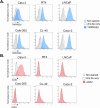
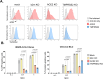
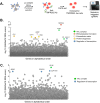
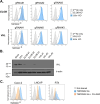
The TMPRSS2 cell surface protease is used by a broad range of respiratory viruses to facilitate entry into target cells. Together with ACE2, TMPRSS2 represents a key factor for SARS-CoV-2 infection, as TMPRSS2 mediates cleavage of viral spike protein, enabling direct fusion of the viral envelope with the host cell membrane. Since the start of the COVID-19 pandemic, TMPRSS2 has gained attention as a therapeutic target for protease inhibitors which would inhibit SARS-CoV-2 infection, but little is known about TMPRSS2 regulation, particularly in cell types physiologically relevant for SARS-CoV-2 infection. Here, we performed an unbiased genome-wide CRISPR-Cas9 library screen, together with a library targeted at epigenetic modifiers and transcriptional regulators, to identify cellular factors that modulate cell surface expression of TMPRSS2 in human colon epithelial cells. We find that endogenous TMPRSS2 is regulated by the Elongin BC-VHL complex and HIF transcription factors. Depletion of Elongin B or treatment of cells with PHD inhibitors resulted in downregulation of TMPRSS2 and inhibition of SARS-CoV-2 infection. We show that TMPRSS2 is still utilised by SARS-CoV-2 Omicron variants for entry into colonic epithelial cells. Our study enhances our understanding of the regulation of endogenous surface TMPRSS2 in cells physiologically relevant to SARS-CoV-2 infection.
Keywords: CRISPR-Cas9 screen; Colon epithelial cells; Coronavirus entry factors; Hypoxic regulation of surface proteins; Transmembrane serine proteases.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures


References
-
- Paoloni-Giacobino, A., Chen, H., Peitsch, M. C., Rossier, C. & Antonarakis, S. E. Cloning of the TMPRSS2 gene, which encodes a novel serine protease with transmembrane, LDLRA, and SRCR domains and maps to 21q22.3. Genomics44, 309–320 (1997). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

