This is a preprint.
Evolving epigenomics of immune cells in type 1 diabetes at single nuclei resolution
- PMID: 40195977
- PMCID: PMC11975021
- DOI: 10.21203/rs.3.rs-5588592/v1
Evolving epigenomics of immune cells in type 1 diabetes at single nuclei resolution
Abstract
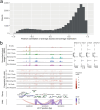
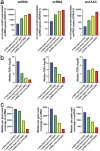
The appearance of diabetes-associated autoantibodies is the first detectable sign of the disease process leading to type 1 diabetes (T1D). Evidence suggests that T1D is a heterogenous disease, where the type of antibodies first formed imply subtypes. Here, we followed 49 children, who subsequently presented with T1D and 49 matched controls, profiling single-cell epigenomics at different time points of disease development. Quantitation of cell and nuclei populations as well as transcriptome and open-chromatin states indicated robust, early, replicable monocyte lineage differences between cases and controls, suggesting heightened pro-inflammatory cytokine secretion early among cases. The order of autoantibody emergence in cases showed variation across lymphoid and myeloid cells, potentially indicating cellular immune response divergence. The strong monocytic lineage representation in peripheral blood immune cells before seroconversion and the weaker differential coordination of these gene networks close to clinical diagnosis emphasizes the importance of early life as a critical phase in T1D development.
Conflict of interest statement
COMPETING INTERESTS The authors declare no competing interests.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

