This is a preprint.
A fragment-based electrophile-first approach to target histidine with aryl-fluorosulfates: application to hMcl-1
- PMID: 40196002
- PMCID: PMC11975029
- DOI: 10.21203/rs.3.rs-6214862/v1
A fragment-based electrophile-first approach to target histidine with aryl-fluorosulfates: application to hMcl-1
Update in
-
A Fragment-Based Electrophile-First Approach to Target Histidine with Aryl-Fluorosulfates: Application to hMcl-1.J Med Chem. 2025 Nov 27;68(22):24305-24315. doi: 10.1021/acs.jmedchem.5c02199. Epub 2025 Nov 12. J Med Chem. 2025. PMID: 41223148 Free PMC article.
Abstract
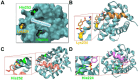
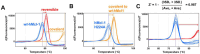
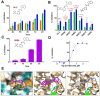
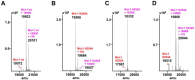
Aryl-fluorosulfates are mild electrophiles that are very stable in biological media and in vivo and can efficiently react with the side chains of Lys, Tyr, or His residues, when properly juxtaposed by a high-affinity ligand. A more powerful approach to derive novel ligands would consist in starting from the covalent adduct and building the ligand off those initial interactions. While this strategy has been proven for Cys with molecular fragments containing Cys targeting electrophiles such as acrylamides, a corresponding strategy with fluorosulfates targeting His/Lys/Tyr residues has yet to be reported. We report here that a fragment library of aryl-fluorosulfates, when deployed with proper biophysical screening strategies, can identify initial covalent fragments. We report on novel strategies to enhance the success rate of such electrophile-based fragment screening for His/Lys/Tyr residues and to characterize the resulting hits. As an application we report on novel covalent fragment hits targeting hMcl-1 His 224.
Keywords: Histidine covalent; aryl-fluorosulfates; drug discovery; electrophile-first; fragment-based drug design; fragment-based ligand design; hMcl-1.
Conflict of interest statement
Conflicts of Interest: GA declares no conflict of interest. MP is co-founder of Armida Labs, Inc. KM and ZA are employees of Cayman Chemical.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

