Evaluation of extended-spectrum β-lactamase producing bacteria in feces of shelter dogs as a biomarker for altered gut microbial taxa and functional profiles
- PMID: 40196031
- PMCID: PMC11975251
- DOI: 10.3389/fmicb.2025.1556442
Evaluation of extended-spectrum β-lactamase producing bacteria in feces of shelter dogs as a biomarker for altered gut microbial taxa and functional profiles
Abstract
Background: The USA is home to 83-88 million dogs, with 3-7 million living in shelters. Shelter dogs move through the supply chain from their geographical origin to adoptive homes, with possible exposure to pathogens and shift in their gut microbiota. However, research in this area is limited. This study examined the effects of intestinal colonization by ESBL bacteria on gut taxa abundance, diversity, and functions in 52 shelter dogs of various ages, sexes, and fertility statuses.
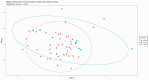
Methodology: We isolated fecal DNA, sequenced their 16S, processed the sequences using DADA2, identified taxa profiles in each dog by Phyloseq, and analyzed Chao1, Shannon, and Simpson alpha diversity by ggplot2 and Wilcoxon test. We analyzed beta diversity using Bray-Curtis dissimilarity matrix from the vegan package. Differential abundance of taxa, gut microbiome functions, and differential abundance of microbiome functions were analyzed using DESeq2, PICRUSt2, and ALDEx2, respectively, with Wilcoxon rank and Kruskal-Wallis tests for comparisons between dog groups.
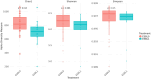
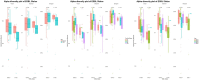
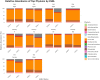
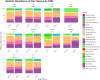
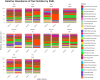
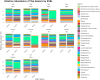
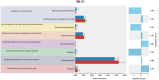
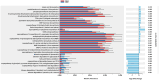
Results: Firmicutes (69.3%), Bacteroidota (13.5%), Actinobacteriota (6.77%), Proteobacteria (5.54%), and Fusobacteriota (4.75%) were the major phyla in the gut of shelter dogs. ESBL bacteria colonized dogs had reduced gut microbiota alpha diversity than non-colonized dogs. The abundance levels of the following phyla (Proteobacteria, Deferribacterota, Bacteroidota, Fusobacteriota, and Spirochaetota), class (Gammaproteobacteria, Bacteroidia, Deferribacteres, Brachyspirae, and Fusobacteria), and families (Enterobacteriaceae, Peptostreptococcaceae, Lactobacillaceae, Lachnospiraceae, Prevotellaceae, and Peptostreptococcaceae) were significantly (p < 0.05) varied between the two dog groups. Further stratified analysis by age, sex, and spaying/neutering status influenced the abundance of taxa in ESBL bacteria colonized dogs, indicating these covariates act as effect modifiers. Most gut metabolic and biosynthetic pathways were downregulated in ESBL bacteria colonized dogs compared to non-colonized dogs. However, alpha-linolenic acid metabolism and shigellosis, fluorobenzoate degradation, allantoin degradation, toluene degradation, glycol degradation, fatty acid and beta-oxidation, and glyoxylate metabolism bypass pathways were increased in dogs colonized by ESBL bacteria.
Conclusion: Colonization by ESBL bacteria marks altered gut microbiota. Dog's demography and fertility status modify the alterations, indicating host factors and ESBL bacteria interplay to shape gut microbiota. ESBL bacteria or other factors reprogram gut microbiome functions through down and upregulating multiple metabolic and biosynthesis pathways to promote ESBL bacteria colonization.
Keywords: 16S amplicon sequencing; ESBL bacteria; alpha diversity; gut microbiota; microbiome function; shelter dogs.
Copyright © 2025 Abdi, Datta, Zawar and Kafle.
Conflict of interest statement
SD and AZ were employed by GeneSpectrum Life Sciences LLP. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Gut Colonization by ESBL-Producing Escherichia coli in Dogs Is Associated with a Distinct Microbiome and Resistome Composition.Microbiol Spectr. 2023 Aug 17;11(4):e0006323. doi: 10.1128/spectrum.00063-23. Epub 2023 Jul 5. Microbiol Spectr. 2023. PMID: 37404183 Free PMC article.
-
Gut microbiome diversity and composition in individuals with and without extended-spectrum β-lactamase-producing Enterobacterales carriage: a matched case-control study in infectious diseases department.Clin Microbiol Infect. 2024 Sep;30(9):1154-1163. doi: 10.1016/j.cmi.2024.03.016. Epub 2024 Mar 23. Clin Microbiol Infect. 2024. PMID: 38527613
-
[Analysis of the dynamic changes in gut microbiota in patients with extremely severe burns by 16S ribosomal RNA high-throughput sequencing technology].Zhonghua Shao Shang Za Zhi. 2020 Dec 20;36(12):1159-1166. doi: 10.3760/cma.j.cn501120-20200518-00271. Zhonghua Shao Shang Za Zhi. 2020. PMID: 33379852 Chinese.
-
Gut microbiome signatures of nursing home residents carrying Enterobacteria producing extended-spectrum β-lactamases.Antimicrob Resist Infect Control. 2020 Jul 14;9(1):107. doi: 10.1186/s13756-020-00773-y. Antimicrob Resist Infect Control. 2020. PMID: 32665016 Free PMC article.
-
Gut microbiota alterations in colorectal adenoma-carcinoma sequence based on 16S rRNA gene sequencing: A systematic review and meta-analysis.Microb Pathog. 2024 Oct;195:106889. doi: 10.1016/j.micpath.2024.106889. Epub 2024 Aug 26. Microb Pathog. 2024. PMID: 39197689
References
-
- Alessandri G., Argentini C., Milani C., Turroni F., Cristina Ossiprandi M., van Sinderen D., et al. . (2020). Catching a glimpse of the bacterial gut community of companion animals: a canine and feline perspective. Microb. Biotechnol. 13, 1708–1732. doi: 10.1111/1751-7915.13656, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

