Multi-trait phenotypic modeling through factor analysis and bayesian network learning to develop latent reproductive, body conformational, and carcass-associated traits in admixed beef heifers
- PMID: 40196222
- PMCID: PMC11973389
- DOI: 10.3389/fgene.2025.1551967
Multi-trait phenotypic modeling through factor analysis and bayesian network learning to develop latent reproductive, body conformational, and carcass-associated traits in admixed beef heifers
Abstract
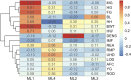
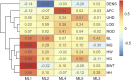
Despite high-throughput and large-scale phenotyping becoming easier, interpretation of such data in cattle production remains challenging due to the complex and highly correlated nature of many traits. Underlying biological traits (UBT) of economic importance are defined by a subset of easy-to-measure traits, leading to challenges in making appropriate selection decisions on them. Research on UBT in beef cattle is limited. In this study, the phenotypic data of admixed beef heifers (n = 336) for reproductive, body conformation, and carcass-related traits (traits, t = 35) were used to identify latent variables from factor analysis (FA) that can be characterized as UBT. Given sample size constraints for carcass (n = 161) and other body size-related traits (n = 336), two models were explored. In Model 1, all individual traits were considered (n = 161), while in Model 2, the dataset was split into body size (n = 336) and carcass (n = 161) traits to maximize available heifers per dataset. A combination of FA and Bayesian network (BN) learning was adopted to develop UBT and infer BN structure for subsequent analyses. All heifers (n = 336) were genotyped using GeneSeek Genomic Profiler 150K for Beef Cattle. Following quality checks, 117,373 autosomal SNP markers were retained and used for genomic estimated breeding values (gEBV) in BN learning steps. Using exploratory and confirmatory FA, Body Size (BS) and Body Composition (BC) were identified as UBT for Model 1, explaining 14 phenotypic traits (t = 14). For Model 2, BS, Ovary Size, and Yield Grade (YG) were identified as UBT, explaining 12 phenotypic traits (t = 12). When using gEBV, the causal network structure inferred showed BS contributed to BC in Model 1 and to Ovary Size in Model 2. Therefore, a structure equation-based approach should be used in subsequent modeling for these traits. From Model 2, YG should be modeled univariately. This study is the first to identify UBT in growing admixed heifers using body size, conformation, and carcass traits. We also identified that BC and YG did not explain intra-muscular fat and body density, indicating these two traits should also be modeled univariately.
Keywords: Bayesian network; factor analytic models; heifer development; latent phenotypes; multi-trait modeling; phenomics.
Copyright © 2025 Anas, Zhao, Yu, Dahlen, Swanson, Ringwall and Hulsman Hanna.
Conflict of interest statement
The authors also declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures





Similar articles
-
Genetic parameters for carcass traits of progeny of beef bulls mated to dairy cows.J Anim Sci. 2024 Jan 3;102:skae075. doi: 10.1093/jas/skae075. J Anim Sci. 2024. PMID: 38489760 Free PMC article.
-
National Beef Quality Audit-2011: In-plant survey of targeted carcass characteristics related to quality, quantity, value, and marketing of fed steers and heifers.J Anim Sci. 2012 Dec;90(13):5143-51. doi: 10.2527/jas.2012-5550. Epub 2012 Sep 5. J Anim Sci. 2012. PMID: 22952369
-
Development of genomic predictions for Angus cattle in Brazil incorporating genotypes from related American sires.J Anim Sci. 2022 Feb 1;100(2):skac009. doi: 10.1093/jas/skac009. J Anim Sci. 2022. PMID: 35031806 Free PMC article.
-
Genomic evaluation of carcass traits of Korean beef cattle Hanwoo using a single-step marker effect model.J Anim Sci. 2023 Jan 3;101:skad104. doi: 10.1093/jas/skad104. J Anim Sci. 2023. PMID: 37004242 Free PMC article.
-
Beef heifer fertility: importance of management practices and technological advancements.J Anim Sci Biotechnol. 2020 Oct 1;11:97. doi: 10.1186/s40104-020-00503-9. eCollection 2020. J Anim Sci Biotechnol. 2020. PMID: 33014361 Free PMC article. Review.
References
-
- Campos R. V., Cobuci J. A., Costa C. N., Braccini Neto J. (2012). Genetic parameters for type traits in Holstein cows in Brazil. Rev. Bras. Zootecn. 41, 2150–2161. 10.1590/s1516-35982012001000003 - DOI
LinkOut - more resources
Full Text Sources

