This is a preprint.
Genomic investigation of MRSA bacteremia relapse reveals diverse genomic profiles but convergence in bacteremia-associated genes
- PMID: 40196254
- PMCID: PMC11974793
- DOI: 10.1101/2025.03.24.25324140
Genomic investigation of MRSA bacteremia relapse reveals diverse genomic profiles but convergence in bacteremia-associated genes
Update in
-
Methicillin-Resistant Staphylococcus aureus Bacteremia Relapses Show Diverse Genomic Profiles but Convergence in Bacteremia-Associated Genes.J Infect Dis. 2025 Dec 20;232(6):1338-1350. doi: 10.1093/infdis/jiaf352. J Infect Dis. 2025. PMID: 40709822
Abstract
Background: Recurrence of methicillin-resistant Staphylococcus aureus (MRSA) bacteremia is a high risk complication for patients. Distinguishing persistent lineages from new infections is not standardized across clinical studies.
Methods: We investigated factors contributing to recurrence of MRSA bacteremia among subjects in Philadelphia, Pennsylvania. Subject demographics and clinical history were collected and paired with whole-genome sequences of infection isolates. Recurrent bacteremia episodes were recorded and defined as relapse infections (same lineage) or new infections by genomic criteria, where a relapse contains isolates <=25 single nucleotide polymorphisms (SNP) different, and by clinical criteria. All isolates were assessed for pairwise SNP distances, common mutations, and signatures of within-host adaptation using the McDonald-Kreitman test. Clusters of transmission between relapse-associated isolates and other subject lineages were identified.
Results: Among 411 sequential subjects with MRSA bacteremia, 32 experienced recurrent bacteremia episodes, with 24 subjects having exclusively relapse infections, six with infections exclusively from a new strain, and two patients with both relapse and new infections. No concordance between a genomic and a clinical definition of relapse was evident (Cohen's Kappa = 0.18, CI: -0.41). Recurrence-associated lineages exhibited signatures of positive selection(G-test:<0.01). Genes with SNPs occurring in multiple relapse lineages have roles in antibiotic resistance and virulence, including 5 lineages with mutations in mprF and 3 lineages with mutations in rpoB, which corresponded with evolved phenotypic changes in daptomycin and rifampin resistance.
Conclusions: Recurrent infections have a diverse strain background. Relapses can be readily distinguished from newly acquired infections using genomic sequencing but not clinical criteria.
Keywords: antibiotic resistance; genomic epidemiology; hospital epidemiology; infection prevention.
Conflict of interest statement
Conflicts of Interest The authors report no conflicts of interest.
Figures
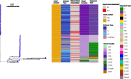




References
-
- Raad I, Narro J, Khan A, Tarrand J, Vartivarian S, Bodey GP. Serious complications of vascular catheter-related Staphylococcus aureus bacteremia in cancer patients. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol. Germany; 1992; 11(8):675–682. - PubMed
-
- Ehni WF, Reller LB. Short-course therapy for catheter-associated Staphylococcus aureus bacteremia. Arch Intern Med. United States; 1989; 149(3):533–536. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
