This is a preprint.
A single high-zinc activation enhancer can control two genes orientated head-to-head in C. elegans
- PMID: 40196504
- PMCID: PMC11974713
- DOI: 10.1101/2024.11.19.624376
A single high-zinc activation enhancer can control two genes orientated head-to-head in C. elegans
Update in
-
A single high-zinc activation enhancer can control two genes oriented head-to-head in Caenorhabditis elegans.G3 (Bethesda). 2025 Jul 9;15(7):jkaf089. doi: 10.1093/g3journal/jkaf089. G3 (Bethesda). 2025. PMID: 40408183 Free PMC article.
Abstract
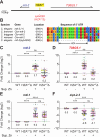
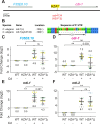
Enhancers play critical roles in gene expression, but a full understanding of their complex functions has yet to be defined. The cellular response to excess zinc levels in C. elegans requires the HIZR-1 transcription factor, which binds the high-zinc activation (HZA) enhancer in the promoters of multiple target genes. Cadmium hijacks the excess zinc response by binding and activating HIZR-1. By analyzing the genome-wide transcriptional response to excess zinc and cadmium, we identified two positions in the genome where head-to-head oriented genes are both induced by metals. In both examples, a single predicted HZA enhancer is positioned between the two translational start sites. We hypothesized that a single enhancer can control both head-to-head genes, an arrangement that has not been extensively characterized. To test this hypothesis, we used CRISPR genome editing to precisely delete the HZAmT enhancer positioned between mtl-2 and T08G5.1; in this mutant, both head-to-head genes display severely reduced zinc-activated transcription, whereas zinc-activated transcription of more distant genes was not strongly affected. Deleting the HZAcF enhancer positioned between cdr-1 and F35E8.10 caused both head-to-head genes to display reduced cadmium-activated transcription, whereas cadmium-activated transcription of more distant genes was not strongly affected. These studies rigorously document that a single HZA enhancer can control two head-to-head genes, advancing our understanding of the diverse functions of enhancers.
Keywords: C. elegans; HZA; Zinc metabolism; divergent transcription; enhancer; high-zinc activation element.
Conflict of interest statement
Conflicts of Interest The authors declare no conflicts of interest.
Figures



Similar articles
-
A single high-zinc activation enhancer can control two genes oriented head-to-head in Caenorhabditis elegans.G3 (Bethesda). 2025 Jul 9;15(7):jkaf089. doi: 10.1093/g3journal/jkaf089. G3 (Bethesda). 2025. PMID: 40408183 Free PMC article.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Assessing the comparative effects of interventions in COPD: a tutorial on network meta-analysis for clinicians.Respir Res. 2024 Dec 21;25(1):438. doi: 10.1186/s12931-024-03056-x. Respir Res. 2024. PMID: 39709425 Free PMC article. Review.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
References
-
- Aggett PJ. 1989. Severe Zinc Deficiency. In: Mills CF, editor. Zinc in Human Biology. London: Springer London. (Macdonald I, editor. ILSI Human Nutrition Reviews). p. 259–279. [accessed 2025 Mar 4]. http://link.springer.com/10.1007/978-1-4471-3879-2_17. - DOI
-
- Almutairi N, Khan N, Harrison-Smith A, Arlt VM, Stürzenbaum SR. 2024. Stage-specific exposure of Caenorhabditis elegans to cadmium identifies unique transcriptomic response cascades and an uncharacterised cadmium responsive transcript. Metallomics.:mfae016. doi: 10.1093/mtomcs/mfae016. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
