This is a preprint.
PTRAMP, CSS and Ripr form a conserved complex required for merozoite invasion of Plasmodium species into erythrocytes
- PMID: 40196582
- PMCID: PMC11974866
- DOI: 10.1101/2025.03.25.644866
PTRAMP, CSS and Ripr form a conserved complex required for merozoite invasion of Plasmodium species into erythrocytes
Update in
-
PTRAMP, CSS and Ripr form a conserved complex required for merozoite invasion of Plasmodium species into erythrocytes.Nat Commun. 2026 Jan 26;17(1):1780. doi: 10.1038/s41467-026-68486-1. Nat Commun. 2026. PMID: 41587959 Free PMC article.
Abstract
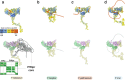
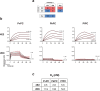
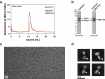
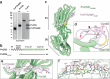
Invasion of erythrocytes by members of the Plasmodium genus is an essential step of the parasite lifecycle, orchestrated by numerous host-parasite interactions. In P. falciparum Rh5, with PfCyRPA, PfRipr, PfCSS, and PfPTRAMP, forms the essential PCRCR complex which binds basigin on the erythrocyte surface. Rh5 is restricted to P. falciparum and its close relatives; however, PTRAMP, CSS and Ripr orthologs are present across the Plasmodium genus. We investigated PTRAMP, CSS and Ripr orthologs from three species to elucidate common features of the complex. Like P. falciparum, PTRAMP and CSS form a disulfide-linked heterodimer in both P. vivax and P. knowlesi with all three species forming a complex (PCR) with Ripr by binding its C-terminal region. Cross-reactive antibodies targeting the PCR complex differentially inhibit merozoite invasion. Cryo-EM visualization of the P. knowlesi PCR complex confirmed predicted models and revealed a core invasion scaffold in Plasmodium spp. with implications for vaccines targeting multiple species of malaria-causing parasites.
Keywords: CSS; P. cynomolgi; P. falciparum; P. knowlesi; P. vivax; PTRAMP; Ripr; erythrocytes; invasion complex; malaria.
Conflict of interest statement
Competing interests The authors have no conflicts of interest to declare.
Figures












References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
