This is a preprint.
Translating the Post-Mortem Brain Multi-Omics Molecular Taxonomy of Alzheimer's Dementia to Living Humans
- PMID: 40196602
- PMCID: PMC11974700
- DOI: 10.1101/2025.03.20.644323
Translating the Post-Mortem Brain Multi-Omics Molecular Taxonomy of Alzheimer's Dementia to Living Humans
Abstract
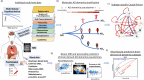
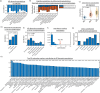
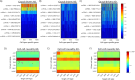
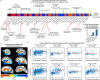
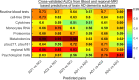
Alzheimer's disease (AD) dementia is characterized by significant molecular and phenotypic heterogeneity, which confounds its mechanistic understanding, diagnosis, and effective treatment. In this study, we harness the most comprehensive dataset of paired ante-mortem blood omics, clinical, psychological, and post-mortem brain multi-omics data and neuroimaging to extensively characterize and translate the molecular taxonomy of AD dementia to living individuals. First, utilizing a comprehensive integration of eight complementary molecular layers from brain multi-omics data (N = 1,189), we identified three distinct molecular AD dementia subtypes exhibiting strong associations with cognitive decline, sex, psychological traits, brain morphology, and characterized by specific cellular and molecular drivers involving immune, vascular, and oligodendrocyte precursor cells. Next, in a significant translational effort, we developed predictive models to convert these advanced brain-derived molecular profiles (AD dementia pseudotimes and subtypes) into blood-, MRI- and psychological traits-based markers. The translation results underscore both the promise of these models and the opportunities for further enhancement. Our findings enhance the understanding of AD heterogeneity, underscore the value of multi-scale molecular approaches for elucidating causal mechanisms, and lay the groundwork for the development of novel therapies in living persons that target multi-level brain molecular subtypes of AD dementia.
Conflict of interest statement
Competing interests: The authors declare no competing interest.
Figures



References
-
- Neff R. A., Wang M., Vatansever S., Guo L., Ming C., Wang Q., Wang E., Horgusluoglu-Moloch E., Song W. M., Li A., Castranio E. L., Julia T. C. W., Ho L., Goate A., Fossati V., Noggle S., Gandy S., Ehrlich M. E., Katsel P., Schadt E., Cai D., Brennand K. J., Haroutunian V., Zhang B., Molecular subtyping of Alzheimer’s disease using RNA sequencing data reveals novel mechanisms and targets, Sci. Adv. 7, 1–18 (2021). - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
