This is a preprint.
Integrated single-cell and spatial analysis identifies context-dependent myeloid-T cell interactions in head and neck cancer immune checkpoint blockade response
- PMID: 40196610
- PMCID: PMC11974738
- DOI: 10.1101/2025.03.24.644582
Integrated single-cell and spatial analysis identifies context-dependent myeloid-T cell interactions in head and neck cancer immune checkpoint blockade response
Abstract
Background: Approximately 15-20% of head and neck cancer squamous cell carcinoma (HNSCC) patients respond favorably to immune checkpoint blockade (ICB). Previous single-cell RNA-Seq (scRNA-Seq) studies identified immune features, including macrophage subset ratios and T-cell subtypes, in HNSCC ICB response. However, the spatial features of HNSCC-infiltrated immune cells in response to ICB treatment need to be better characterized.
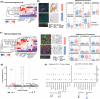
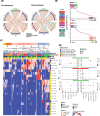
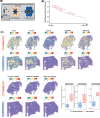
Methods: Here, we perform a systematic evaluation of cell interactions between immune cell types within the tumor microenvironment using spatial omics data using complementary techniques from both 10X Visium spot-based spatial transcriptomics and Nanostring CosMx single-cell spatial omics with RNA gene panel including 435 ligands and receptors. In this study, we used integrated bioinformatics analyses to identify cellular neighborhoods of co-localizing cell types in single-cell spatial transcriptomics and proteomics data. In addition, we used both publicly available scRNA-Seq and in-house spatial RNA-Seq data to identify spatially constrained Ligand-Receptor interactions in Responder patients.
Results: With 522,399 single cells profiled with both RNA and protein from 26 patients, in addition to spot-resolved spatial RNA-Seq from 8 patients treated with ICB together with bioinformatics analysis of publicly available single-cell and bulk RNA-Seq, we have identified a spatial and cell-type specific context-dependency of myeloid and T cell interaction difference between Responders and Non-Responders. We defined further cellular neighborhood and the sources of chemokine CXCL9/10-CXCR3 interactions in Responders, emerging targets in ICB, as well as CXCL16-CXCR6, CCL4/5-CCR5, and other underappreciated and potential markers and targets for ICB response in HNSCC. In addition, we have contributed a rich data resource of cell-cell Ligand Receptor interactions for the immunotherapy and HNSCC research community.
Discussion: Our work provides a comprehensive single-cell and spatial atlas of immune cell interactions that correlate with response to ICB in HNSCC. We showcase how integrating multiple technologies and bioinformatics approaches can provide new insights into potential immune-based biomarkers of ICB response. Our results suggested refining future studies using preclinical animal models in a more context-specific manner to elucidate potential underlying mechanisms that lead to improved ICB responses.
Keywords: cell-cell interactions; cellular neighborhoods; head and neck squamous cell carcinoma; immune checkpoint blockade; ligand-receptor interactions; myeloid cells; scRNA-seq; spatial transcriptomics.
Conflict of interest statement
Competing interests All authors have confirmed no competing interests.
Figures



References
-
- Burtness B. et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): a randomised, open-label, phase 3 study. The Lancet 394, 1915–1928 (2019). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
