This is a preprint.
A factor-based analysis of individual human microglia uncovers regulators of an Alzheimer-related transcriptional signature
- PMID: 40196633
- PMCID: PMC11974870
- DOI: 10.1101/2025.03.27.641500
A factor-based analysis of individual human microglia uncovers regulators of an Alzheimer-related transcriptional signature
Abstract
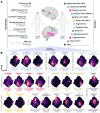
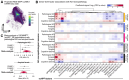
Human microglial heterogeneity is only beginning to be appreciated at the molecular level. Here, we present a large, single-cell atlas of expression signatures from 441,088 live microglia broadly sampled across a diverse set of brain regions and neurodegenerative and neuroinflammatory diseases obtained from 161 donors sampled at autopsy or during a neurosurgical procedure. Using single-cell hierarchical Poisson factorization (scHPF), we derived a 23-factor model for continuous gene expression signatures across microglia which capture specific biological processes (e.g., metabolism, phagocytosis, antigen presentation, inflammatory signaling, disease-associated states). Using external datasets, we evaluated the aspects of microglial phenotypes that are encapsulated in various in vitro and in vivo microglia models and identified and replicated the role of two factors in human postmortem tissue of Alzheimer's disease (AD). Further, we derived a complex network of transcriptional regulators for all factors, including regulators of an AD-related factor enriched for the mouse disease-associated microglia 2 (DAM2) signature: ARID5B, CEBPA, MITF, and PPARG. We replicated the role of these four regulators in the AD-related factor and then designed a multiplexed MERFISH panel to assess our microglial factors using spatial transcriptomics. We find that, unlike cells with high expression of the interferon-response factor, cells with high expression of the AD DAM2-like factor are widely distributed in neocortical tissue. We thus propose a novel analytic framework that provides a taxonomic approach for microglia that is more biologically interpretable and use it to uncover new therapeutic targets for AD.
Figures



References
-
- Li Q. & Barres B. A. Microglia and macrophages in brain homeostasis and disease. Nat. Rev. Immunol. 18, 225–242 (2018). - PubMed
-
- Mittelbronn M., Dietz K., Schluesener H. J. & Meyermann R. Local distribution of microglia in the normal adult human central nervous system differs by up to one order of magnitude. Acta Neuropathol. (Berl.) 101, 249–255 (2001). - PubMed
Publication types
Grants and funding
- P30 AG072975/AG/NIA NIH HHS/United States
- U01 AG046152/AG/NIA NIH HHS/United States
- R01 NS103473/NS/NINDS NIH HHS/United States
- U01 AG061356/AG/NIA NIH HHS/United States
- R01 AG017917/AG/NIA NIH HHS/United States
- P30 AG066462/AG/NIA NIH HHS/United States
- P30 AG010161/AG/NIA NIH HHS/United States
- U01 AG006781/AG/NIA NIH HHS/United States
- RF1 AG057473/AG/NIA NIH HHS/United States
- R01 AG048015/AG/NIA NIH HHS/United States
- R01 AG070438/AG/NIA NIH HHS/United States
- U19 AG066567/AG/NIA NIH HHS/United States
- R01 AG015819/AG/NIA NIH HHS/United States
- F30 CA261090/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
