This is a preprint.
X-Linked Cancer-Associated Polypeptide (XCP) from lncRNA1456 Cooperates with PHF8 to Regulate Gene Expression and Cellular Pathways in Breast Cancer
- PMID: 40196671
- PMCID: PMC11974697
- DOI: 10.1101/2025.03.21.644649
X-Linked Cancer-Associated Polypeptide (XCP) from lncRNA1456 Cooperates with PHF8 to Regulate Gene Expression and Cellular Pathways in Breast Cancer
Abstract
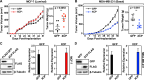
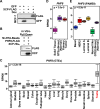
Recent studies have demonstrated that a subset of long "noncoding" RNAs (lncRNAs) produce functional polypeptides and proteins. In this study, we discovered a 132 amino acid protein in human breast cancer cells named XCP (X-linked Cancer-associated Polypeptide), which is encoded by lncRNA1456 (a.k.a. RHOXF1P3), a transcript previously thought to be noncoding. lncRNA1456 is a pancreas- and testis-specific RNA whose gene is located on chromosome X. We found that the expression of lncRNA1456 and XCP are highly upregulated in the luminal A, luminal B, and HER2 molecular subtypes of breast cancer. XCP modulates both estrogen-dependent and estrogen-independent growth of breast cancer cells by regulating cancer pathways, as shown in cell and xenograft models. XCP shares some homology with homeodomain-containing proteins and interacts with the histone demethylase plant homeodomain finger protein 8 (PHF8), which is also encoded by an X-linked gene. Mechanistically, XCP stimulates the histone demethylase activity of PHF8 to regulate gene expression in breast cancer cells. These findings identify XCP as a coregulator of transcription and emphasize the need to interrogate the potential functional roles of open reading frames originating from noncoding RNAs.
Keywords: PHF8; X-Linked Cancer-Associated Polypeptide (XCP); breast cancer; cancer cell growth; chromatin; histone demethylase; lncRNA.
Conflict of interest statement
Declaration of interests The authors have no competing interests to declare.
Figures





Similar articles
-
Histone demethylase PHF8 regulates hypoxia signaling through HIF1α and H3K4me3.Biochim Biophys Acta Gene Regul Mech. 2017 Sep;1860(9):1002-1012. doi: 10.1016/j.bbagrm.2017.07.005. Epub 2017 Jul 20. Biochim Biophys Acta Gene Regul Mech. 2017. PMID: 28734980 Free PMC article.
-
The X-linked mental retardation gene PHF8 is a histone demethylase involved in neuronal differentiation.Cell Res. 2010 Aug;20(8):908-18. doi: 10.1038/cr.2010.81. Epub 2010 Jun 15. Cell Res. 2010. PMID: 20548336
-
Histone demethylase PHF8 promotes progression and metastasis of gastric cancer.Am J Cancer Res. 2017 Mar 1;7(3):448-461. eCollection 2017. Am J Cancer Res. 2017. PMID: 28401003 Free PMC article.
-
Long non-coding RNAs: From disease code to drug role.Acta Pharm Sin B. 2021 Feb;11(2):340-354. doi: 10.1016/j.apsb.2020.10.001. Epub 2020 Oct 10. Acta Pharm Sin B. 2021. PMID: 33643816 Free PMC article. Review.
-
PHF8/KDM7B: A Versatile Histone Demethylase and Epigenetic Modifier in Nervous System Disease and Cancers.Epigenomes. 2024 Sep 15;8(3):36. doi: 10.3390/epigenomes8030036. Epigenomes. 2024. PMID: 39311138 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
