This is a preprint.
Studies on Gene Enhancer with KSHV mini-chromatin
- PMID: 40196677
- PMCID: PMC11974746
- DOI: 10.1101/2025.03.24.644916
Studies on Gene Enhancer with KSHV mini-chromatin
Abstract
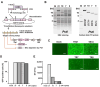
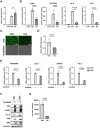
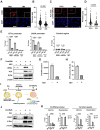
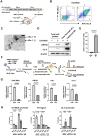
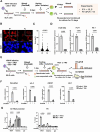
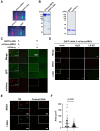
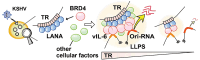
Kaposi's sarcoma-associated herpesvirus (KSHV) genome contains a terminal repeats (TR) sequence. Previous studies demonstrated that KSHV TR functions as a gene enhancer for inducible lytic gene promoters. Gene enhancers anchor bromodomain-containing protein 4 (BRD4) at specific genomic region, where BRD4 interacts flexibly with transcription-related proteins through its intrinsically disordered domain and exerts transcription regulatory function. Here, we generated recombinant KSHV with reduced TR copy numbers and studied BRD4 recruitment and its contributions to the inducible promoter activation. Reducing the TR copy numbers from 21 (TR21) to 5 (TR5) strongly attenuated viral gene expression during de novo infection and impaired reactivation. The EF1α promoter encoded in the KSHV BAC backbone also showed reduced promoter activity, suggesting a global attenuation of transcription activity within TR5 latent episomes. Isolation of reactivating cells confirmed that the reduced inducible gene transcription from TR-shortened DNA template and is mediated by decreased efficacies of BRD4 recruitment to viral gene promoters. Separating the reactivating iSLK cell population from non-responders showed that reactivatable iSLK cells harbored larger LANA nuclear bodies (NBs) compared to non-responders. The cells with larger LANA NBs, either due to prior transcription activation or TR copy number, supported KSHV reactivation more efficiently than those with smaller LANA NBs. With auxin-inducible LANA degradation, we confirmed that LANA is responsible for BRD4 occupancies on latent chromatin. Finally, with purified fluorescence-tagged proteins, we demonstrated that BRD4 is required for LANA to form liquid-liquid phase-separated dots. The inclusion of TR DNA fragments further facilitated the formation of larger BRD4-containing LLPS in the presence of LANA, similar to the "cellular enhancer dot" formed by transcription factor-DNA bindings. These results suggest that LANA binding to TR establishes an enhancer domain for infected KSHV episomes. The strength of this enhancer, regulated by TR length or transcription memory, determines the outcome of KSHV replication.
Conflict of interest statement
Competing interests: YI declares a competing interest relating to founding role for VGN Bio Inc. All other authors declare they have no competing interests.
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
