Altered precipitation and nighttime warming reshape the vertical distribution of soil microbial communities
- PMID: 40197052
- PMCID: PMC12090752
- DOI: 10.1128/msystems.01248-24
Altered precipitation and nighttime warming reshape the vertical distribution of soil microbial communities
Abstract
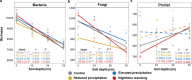
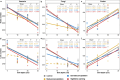
Soil depth determines microbial community composition. Yet, it remains largely unexplored how climate changes affect the vertical distribution of soil microbial communities. Here, we investigated the effects of altered precipitation and nighttime warming on microbial communities in the topsoils (0-20 cm) and subsoils (20-50 cm) of a temperate grassland in Inner Mongolia, China. As commonly observed under nutrient scarcity conditions, bacterial and fungal α-diversity and network complexity decreased with soil depth. However, protistan α-diversity and network complexity increased, which was attributed to less niche overlap and smaller body size. Strikingly, the slopes of linear regressions of microbial α-diversity/network complexity and soil depth were all reduced by altered precipitation. Microbial community composition was significantly influenced by both depth and reduced precipitation, and to a lesser extent by nighttime warming and elevated precipitation. The ribosomal RNA gene operon (rrn) copy number, a genomic proxy of bacterial nutrient demand, decreased with soil depth, and the percentages of positive network links were higher in the subsoil, supporting the "hunger game" hypothesis. Both reduced precipitation and nighttime warming decreased the rrn copy number in the subsoils while increasing the percentages of positive links, enhancing potential niche sharing among bacterial species. The stochasticity level of bacterial and fungal community assemblies decreased with soil depth, showing that depth acted as a selection force. Altered precipitation increased stochasticity, attenuating the depth's filtering effect and diminishing its linear relationship with microbial diversity. Collectively, we unveiled the predominant influence of altered precipitation in affecting the vertical distribution of soil microbial communities.IMPORTANCEUnderstanding how climate change impacts the vertical distribution of soil microbial communities is critical for predicting ecosystem responses to global environmental shifts. Soil microbial communities exhibit strong depth-related stratification, yet the effects of climate change variables, such as altered precipitation and nighttime warming, on these vertical patterns have been inadequately studied. Our research uncovers that altered precipitation disrupts the previously observed relationships between soil depth and microbial diversity, a finding that challenges traditional models of soil microbial ecology. Furthermore, our study provides experimental support for the hunger game hypothesis, highlighting that oligotrophic microbes, characterized by lower ribosomal RNA gene operon (rrn) copy numbers, are selectively favored in nutrient-poor subsoils, fostering increased microbial cooperation for resource exchange. By unraveling these complexities in soil microbial communities, our findings offer crucial insights for predicting ecosystem responses to climate change and for developing strategies to mitigate its adverse impacts.
Keywords: climate change; microbial diversity; microbial network; protistan community; vertical spatial distribution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Stocker T, Plattner G-K, Dahe Q. The physical science basis - findings and lessons learned, p 17003
-
- Cardinale BJ, Duffy JE, Gonzalez A, Hooper DU, Perrings C, Venail P, Narwani A, Mace GM, Tilman D, Wardle DA, Kinzig AP, Daily GC, Loreau M, Grace JB, Larigauderie A, Srivastava DS, Naeem S. 2012. Biodiversity loss and its impact on humanity. Nature 486:59–67. doi: 10.1038/nature11148 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
