Rickettsia rickettsii RoaM negatively regulates expression of a limited number of rickettsial genes
- PMID: 40197162
- PMCID: PMC12039225
- DOI: 10.1128/msphere.00077-25
Rickettsia rickettsii RoaM negatively regulates expression of a limited number of rickettsial genes
Abstract
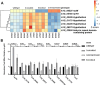
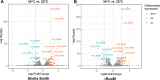
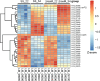
The recently described rickettsial protein RoaM (regulator of actin-based motility) negatively regulates the production of actin tails, and its abrogation induces hyper-spreading behavior in many laboratory-adapted strains of Rickettsia rickettsii. RoaM is not surface exposed; thus, its mechanism of regulating actin-based motility is unclear. Using R. rickettsii strains derived from the virulent Sheila Smith strain that express varying levels of roaM, an RNA-seq experiment was performed. We found that roaM-overexpressing strains downregulate expression of at least six genes which may link the regulatory effects of RoaM to the phenotypic effect on motility. Genes regulated by RoaM were confirmed by RT-qPCR. Among the genes regulated is the secreted effector RarP2, which disrupts the trans-Golgi network. Two of the hypothetical proteins were shown to be secreted via fusion to a glycogen synthase kinase tag, which when phosphorylated reveals exposure to the host-cell cytosol. Taken together, these data support the hypothesis that RoaM affects transcription, downregulating rickettsial genes important for pathogenicity in the mammalian host but which are perhaps otherwise detrimental within the tick vector. To determine how RoaM activity may itself be regulated, we investigated a role of temperature in roaM transcription. RoaM expression itself is not temperature dependent, but many other rickettsial genes are, including some also regulated by RoaM. This suggests that rickettsiae utilize multiple mechanisms to control gene expression in response to environmental signals.
Importance: RoaM was previously shown to repress the production of actin tails by unknown mechanisms. The roaM gene is negatively selected for in cell culture resulting in hyper-spreading mutants. This work reveals that rather than specifically regulating motility in Rickettsia rickettsii, a set of rickettsial genes is downregulated that includes the type IV secreted effector, rarP2, as well as two other secreted, putative effectors. Relatively few secreted effectors have been identified in Rickettsia. RoaM appears to be part of a larger biological program encompassing active spreading in mammalian cells and may be a critical component for R. rickettsii to transition from arthropod to mammalian host.
Keywords: Rickettsia; environmental signaling; gene expression; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Regulator of Actin-Based Motility (RoaM) Downregulates Actin Tail Formation by Rickettsia rickettsii and Is Negatively Selected in Mammalian Cell Culture.mBio. 2022 Apr 26;13(2):e0035322. doi: 10.1128/mbio.00353-22. Epub 2022 Mar 14. mBio. 2022. PMID: 35285700 Free PMC article.
-
Rickettsia rickettsii virulence determinants RARP2 and RapL mitigate IFN-β signaling in primary human dermal microvascular endothelial cells.mBio. 2024 Apr 10;15(4):e0345023. doi: 10.1128/mbio.03450-23. Epub 2024 Mar 6. mBio. 2024. PMID: 38445878 Free PMC article.
-
The Rickettsial Ankyrin Repeat Protein 2 Is a Type IV Secreted Effector That Associates with the Endoplasmic Reticulum.mBio. 2018 Jun 26;9(3):e00975-18. doi: 10.1128/mBio.00975-18. mBio. 2018. PMID: 29946049 Free PMC article.
-
Genetics of rickettsiae.Eur J Epidemiol. 1991 May;7(3):213-21. doi: 10.1007/BF00145669. Eur J Epidemiol. 1991. PMID: 1679397 Review.
-
Progress in rickettsial genome analysis from pioneering of Rickettsia prowazekii to the recent Rickettsia typhi.Ann N Y Acad Sci. 2005 Dec;1063:13-25. doi: 10.1196/annals.1355.003. Ann N Y Acad Sci. 2005. PMID: 16481486 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
