Early treatment and PD1 inhibition enhance HIV-specific functionality of follicular CD8+ T cells
- PMID: 40197363
- PMCID: PMC11981630
- DOI: 10.1172/jci.insight.180309
Early treatment and PD1 inhibition enhance HIV-specific functionality of follicular CD8+ T cells
Abstract
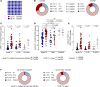
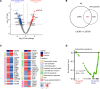
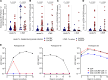
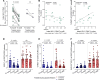
People living with HIV treated during acute infection are the group for whom achieving functional cure appears most viable. Follicular CD8+ T cells could contribute to HIV reservoir clearance by accessing B cell follicles through CXCR5 expression. This study examines peripheral follicular CD8+ T cells using flow cytometry, transcriptome analyses, and functional assays in people treated during acute (n = 37) and chronic (n = 18) infection, as well as in individuals naturally controlling HIV (n = 20) and living without HIV (n = 10). Our results reveal that early, as opposed to late, treatment initiation preserves antiviral effector functions of follicular CD8+ T cells, which are further enhanced by PD1 inhibition. We also identify a correlation between follicular CD8+ T cells and intact proviral HIV DNA levels in acute, but not chronic, infection. Longitudinal transcriptomic analysis of peripheral effector cells after 48 weeks of suppressive therapy indicated traits of recent antigen exposure, suggesting potential recirculation into lymphoid tissue. These findings underscore the pivotal role of follicular CD8+ T cells in anti-HIV responses and support investigating targeted cure strategies, such as anti-PD1 therapy, especially in individuals initiating treatment during acute infection.
Keywords: AIDS/HIV; Adaptive immunity; Immunology; T cells.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

