ATP13A2 as a prognostic biomarker and its correlation with immune infiltration in cervical cancer: A retrospective study
- PMID: 40197818
- PMCID: PMC11976316
- DOI: 10.1111/jcmm.70097
ATP13A2 as a prognostic biomarker and its correlation with immune infiltration in cervical cancer: A retrospective study
Abstract
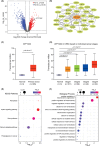
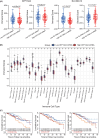
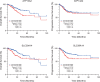
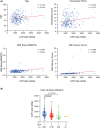
While the oncogene ATP13A2 is reportedly involved in colorectal cancer, its role in cervical cancer (CC) has yet to be fully characterized. In this study, we investigated ATP13A2 as a potential prognostic biomarker of CC. To this end, we compared CC tissues with normal tissues to identify differentially expressed genes, identifying ATP13A2 as a potential marker of CC. Elevated ATP13A2 expression levels were identified in CC samples compared to noncancerous samples across various data sets, with further immunohistochemical validation. Functional enrichment analysis revealed that ATP13A2 plays an essential role in the CXCL12-activated CXCR4 signalling pathway and chemotaxis regulation, which may alter immune infiltration. Notably, increased ATP13A2 levels were associated with poor overall survival. Furthermore, multiple clinical characteristics were significantly associated with ATP13A2 expression. Additionally, tumour bacterial infiltration was assessed using weighted co-expression network analysis, revealing a relationship between ATP13A2 expression and bacteria in the CC tumour microenvironment. Our results suggest that ATP13A2 is a promising diagnostic and prognostic marker for CC. However, further large-scale studies are needed to fully elucidate the mechanisms underlying the involvement of ATP13A2 in CC.
Keywords: ATP13A2; cervical cancer; immune infiltration; microbiome; prognostic marker.
© 2025 The Author(s). Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Identification of immune-related cervical cancer prognostic biomarkers and construction of prognostic model based on tumor microenvironment.Eur J Med Res. 2025 Apr 9;30(1):261. doi: 10.1186/s40001-025-02515-5. Eur J Med Res. 2025. PMID: 40205481 Free PMC article.
-
Ferroptosis-related gene transferrin receptor protein 1 expression correlates with the prognosis and tumor immune microenvironment in cervical cancer.PeerJ. 2024 Aug 6;12:e17842. doi: 10.7717/peerj.17842. eCollection 2024. PeerJ. 2024. PMID: 39131609 Free PMC article.
-
Elucidating the role of tumor-associated ALOX5+ mast cells with transformative function in cervical cancer progression via single-cell RNA sequencing.Front Immunol. 2024 Aug 19;15:1434450. doi: 10.3389/fimmu.2024.1434450. eCollection 2024. Front Immunol. 2024. PMID: 39224598 Free PMC article.
-
CXCR4: a key receptor in the crosstalk between tumor cells and their microenvironment.Blood. 2006 Mar 1;107(5):1761-7. doi: 10.1182/blood-2005-08-3182. Epub 2005 Nov 3. Blood. 2006. PMID: 16269611 Review.
-
Understanding Cervical Cancer through Proteomics.Cells. 2021 Jul 22;10(8):1854. doi: 10.3390/cells10081854. Cells. 2021. PMID: 34440623 Free PMC article. Review.

