Preadipocyte IL-13/IL-13Rα1 signaling regulates beige adipogenesis through modulation of PPARγ activity
- PMID: 40198135
- PMCID: PMC12126228
- DOI: 10.1172/JCI169152
Preadipocyte IL-13/IL-13Rα1 signaling regulates beige adipogenesis through modulation of PPARγ activity
Abstract
Type 2 innate lymphoid cells (ILC2s) regulate the proliferation of preadipocytes that give rise to beige adipocytes. Whether and how ILC2 downstream Th2 cytokines control beige adipogenesis remain unclear. We used cell systems and genetic models to examine the mechanism through which IL-13, an ILC2-derived Th2 cytokine, controls beige adipocyte differentiation. IL-13 priming in preadipocytes drove beige adipogenesis by upregulating beige-promoting metabolic programs, including mitochondrial oxidative metabolism and PPARγ-related pathways. The latter was mediated by increased expression and activity of PPARγ through the IL-13 receptor 1 (IL-13R1) downstream effectors STAT6 and p38 MAPK, respectively. Il13-KO or preadipocyte Il13ra1-KO mice were refractory to cold- or β3-adrenergic agonist-induced beiging in inguinal white adipose tissue, whereas Il4-KO mice showed no defects in beige adipogenesis. Il13-KO and Il13ra1-KO mouse models exhibited increased body weight and fat mass and dysregulated glucose metabolism but had a mild cold-intolerant phenotype, likely due to their intact brown adipocyte recruitment. We also found that genetic variants of human IL13RA1 were associated with BMI and type 2 diabetes. These results suggest that IL-13 signaling-regulated beige adipocyte function may play a predominant role in modulating metabolic homeostasis rather than in thermoregulation.
Keywords: Adipose tissue; Cell biology; Glucose metabolism; Metabolism; Obesity.
Figures
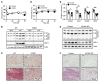


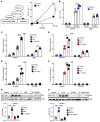
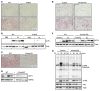
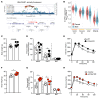
Comment in
- IL-13 priming in precursors drives beige adipogenesis and enhances metabolic homeostasis doi: 10.1172/JCI191361
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

