Generation of induced alveolar assembloids with functional alveolar-like macrophages
- PMID: 40199883
- PMCID: PMC11978882
- DOI: 10.1038/s41467-025-58450-w
Generation of induced alveolar assembloids with functional alveolar-like macrophages
Abstract
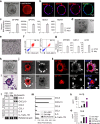
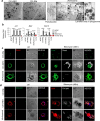
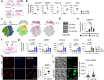
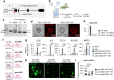
Within the human lung, interactions between alveolar epithelial cells and resident macrophages shape lung development and function in both health and disease. To study these processes, we develop a co-culture system combining human pluripotent stem cell-derived alveolar epithelial organoids and induced macrophages to create a functional environment, termed induced alveolar assembloids. Using single-cell RNA sequencing and functional analyses, we identify alveolar type 2-like cells producing GM-CSF, which supports macrophage tissue adaptation, and macrophage-like cells that secrete interleukin-1β and interleukin-6, express surfactant metabolism genes, and demonstrate core immune functions. In response to alveolar epithelial injury, macrophage-like cells efficiently eliminate damaged cells and absorb oxidized lipids. Exposure to bacterial components or infection with Mycobacterium tuberculosis reveals that these assembloids replicate key aspects of human respiratory defense. These findings highlight the potential of induced alveolar assembloids as a platform to investigate human lung development, immunity, and disease.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
- KGM4722533/Korea Research Institute of Bioscience and Biotechnology (KRIBB)
- 2022M3A9J1072296, RS-2023-00225239, 2021R1C1C1006725/National Research Foundation of Korea (NRF)
- RS-2024-00458236/National Research Foundation of Korea (NRF)
- 2023-NS-001-00/Ministry of Health, Welfare and Family Affairs | Korea Centers for Disease Control & Prevention (KCDC)
LinkOut - more resources
Full Text Sources

