Transcriptomics-driven exploration of genetic variation and peptide discovery in the sea anemones Anthopleura midori and Actinia equina
- PMID: 40200035
- PMCID: PMC11978773
- DOI: 10.1038/s41598-025-96976-7
Transcriptomics-driven exploration of genetic variation and peptide discovery in the sea anemones Anthopleura midori and Actinia equina
Abstract
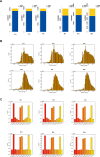
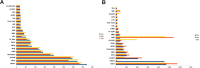
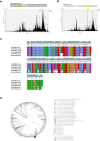
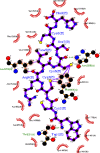
Exploring sea anemone polypeptides enables us to understand the evolutionary history and ecological adaptation strategies of species at the microscopic level. More importantly, it aims to provide a solid theoretical foundation for drug development and biodiversity conservation research. Through systematic research, we discovered a total of 51 toxin sequences in species Anthopleura midori and Acyinia equina. The toxin sequences between the two species exhibited significant differences, with notable diversity observed among individuals. In terms of genetic diversity, species Anthopleura midori primarily exhibits variations due to single nucleotide polymorphisms (SNPs), whereas species Actinia equina shows frequent insertion and deletion events. In transcription factor analysis, both species Anthopleura midori and Actinia equina share common transcription factors TEA (TEA Domain Transcription Factor), SPL(Squamosa Promoter Binding Protein-like), and bHLH (Basic Helix-Loop-Helix). Notably. Notably, bHLH is highly expressed in Actinia equina, which may give it advantages in muscle and nervous system development. On the other hand, Anthopleura midori may rely on other transcription factors. Furthermore, by employing transcriptomics and mass spectrometry techniques, two new gene families were successfully identified, and five structurally novel cyclic peptides were predicted. Kinetic simulations further confirmed that the peptide segment B3a-c29555_c4_g4 binds primarily through hydrogen bonds and hydrophobic interactions with the Cav3.1 (PDB ID:6 KZO) protein, and this peptide has the potential to act as a channel modulator for Cav3.1. Overall, this research not only deepens our understanding of the genetic basis of toxin diversity but also highlights the great potential of these toxins in the development of novel drugs.
Keywords: SNPs; Sea anemones; Toxins; Transcription factors.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures



References
-
- Sigwart, J. D., Blasiak, R., Jaspars, M., Jouffray, J. B. & Tasdemir, D. Unlocking the potential of marine biodiscovery. Nat. Prod. Rep.38, 1235–1242. 10.1039/d0np00067a (2021). - PubMed
-
- Fu, X. Y. et al. Three rounds of Stability-Guided optimization and systematical evaluation of oncolytic peptide LTX-315. J. Med. Chem.67, 3885–3908. 10.1021/acs.jmedchem.3c02232 (2024). - PubMed
-
- Norton, R. S. & Chandy, K. G. Venom-derived peptide inhibitors of voltage-gated potassium channels. Neuropharmacology127, 124–138. 10.1016/j.neuropharm.2017.07.002 (2017). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

