Mechanisms of Baicalin Alleviates Intestinal Inflammation: Role of M1 Macrophage Polarization and Lactobacillus amylovorus
- PMID: 40200426
- PMCID: PMC12140389
- DOI: 10.1002/advs.202415948
Mechanisms of Baicalin Alleviates Intestinal Inflammation: Role of M1 Macrophage Polarization and Lactobacillus amylovorus
Abstract
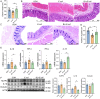
Baicalin has been widely used for its anti-inflammatory pharmacological properties, yet its effects on bacterial intestinal inflammation and the mechanisms remain unclear. This study revealed that baicalin alleviates bacterial intestinal inflammation through regulating macrophage polarization and increasing Lactobacillus amylovorus abundance in colon. Specifically, transcriptomic analysis showed that baicalin restored Escherichia coli-induced genes expression changes including T helper cell 17 differentiation-related genes, macrophage polarization related genes, and TLR/IRF/STAT signaling pathway. Subsequent microbial and non-targeted metabolomic analysis revealed that these changes may be related to the enhancement of Lactobacillus amylovorus and the upregulation of its metabolites including chrysin, lactic acid, and indoles. Furthermore, whole-genome sequencing of Lactobacillus amylovorus provided insights into its functional potential and metabolic annotations. Lactobacillus amylovorus supplementation alleviates Escherichia coli-induced intestinal inflammation in mice and similarly inhibited M1 macrophage polarization through TLR4/IRF/STAT pathway. Additionally, baicalin, Lactobacillus amylovorus, or chrysin alone could regulate macrophage polarization, highlighting their independent anti-inflammatory potential. Notably, this study revealed that baicalin alleviates intestinal inflammation through TLR4/IRF/STAT pathway and increasing Lactobacillus amylovorus abundance and the synthesis of chrysin. These findings provide new insights into the therapeutic potential of baicalin and Lactobacillus amylovorus in preventing and treating intestinal inflammation, offering key targets for future interventions.
Keywords: E. coli; Lactobacillus amylovorus; TLR4; baicalin; intestinal inflammation; macrophages polarization.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- W. H. Organization , Diarrhoeal disease, https://www.who.int/news‐room/fact‐sheets/detail/diarrhoeal‐disease (accessed: March 2024).
-
- Liu L., Oza S., Hogan D., Perin J., Rudan I., Lawn J. E., Cousens S., Mathers C., Black R. E., Lancet 2015, 385, 430. - PubMed
-
- Gan Z., Zhao M., Xia Y., Yan Y., Ren W., Trends Endocrinol. Metab. 2024, 35, 62. - PubMed
-
- Siebeler R., de Winther M. P. J., Hoeksema M. A., J. Allergy Clin. Immunol. 2023, 152, 326. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
