Allele-specific silencing of a dominant SETX mutation in familial amyotrophic lateral sclerosis type 4
- PMID: 40200577
- PMCID: PMC12023877
- DOI: 10.1016/j.xhgg.2025.100435
Allele-specific silencing of a dominant SETX mutation in familial amyotrophic lateral sclerosis type 4
Abstract
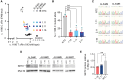
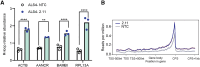
Amyotrophic lateral sclerosis 4 (ALS4) is an autosomal dominant motor neuron disease that is molecularly characterized by reduced R-loop levels and caused by pathogenic variants in senataxin (SETX). SETX encodes an RNA/DNA helicase that resolves three-stranded nucleic acid structures called R-loops. Currently, there are no disease-modifying therapies available for ALS4. Given that SETX is haplosufficient, removing the product of the mutated allele presents a potential therapeutic strategy. We designed a series of siRNAs to selectively target the RNA transcript from the ALS4 allele containing the c.1166T>C mutation (p.Leu389Ser). Transfection of HEK293 cells with siRNA and plasmids encoding either wild-type or mutant (Leu389Ser) epitope-tagged SETX revealed that three siRNAs specifically reduced mutant SETX protein levels while having minimal effect on the wild-type SETX protein. In ALS4 primary fibroblasts, siRNA treatment silenced the endogenous mutant SETX allele while sparing the wild-type allele and restored R-loop levels in patient cells. Our findings demonstrate that mutant SETX, differing from wild-type by a single nucleotide, can be effectively and specifically silenced by RNA interference.
Keywords: amyotrophic lateral sclerosis; motor neuron disease; senataxin; siRNA.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

Update of
-
Allele-specific silencing of a dominant SETX mutation in familial amyotrophic lateral sclerosis type 4.bioRxiv [Preprint]. 2024 Oct 12:2024.10.11.617871. doi: 10.1101/2024.10.11.617871. bioRxiv. 2024. Update in: HGG Adv. 2025 Jul 10;6(3):100435. doi: 10.1016/j.xhgg.2025.100435. PMID: 39416141 Free PMC article. Updated. Preprint.
References
-
- Rabin B.A., Griffin J.W., Crain B.J., Scavina M., Chance P.F., Cornblath D.R. Autosomal dominant juvenile amyotrophic lateral sclerosis. Brain. 1999;122:1539–1550. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

