A competition network connects Rab5 and Rab11 GTPases at the surface of endocytic structures
- PMID: 40201129
- PMCID: PMC11978354
- DOI: 10.1016/j.isci.2025.112170
A competition network connects Rab5 and Rab11 GTPases at the surface of endocytic structures
Abstract
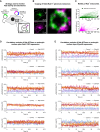
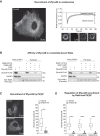
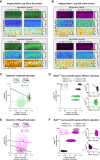
Specificity in membrane trafficking relies on the interaction between Rab small GTPase proteins and their molecular effectors. However, the evidence that different Rab proteins can bind to common effectors challenges this view. Here, we show that molecular competition between distinct Rab GTPases for a shared protein can link diverse membrane trafficking pathways. Theoretical analysis and experimental data support a role for Zfyve26 as a part of a competitive network that modulates changes in Rab5-Rab11 abundance, activation status, and correlation at the surface of single endocytic structures. By leveraging on the Loop index, a novel metric that couples the GTP-bound fraction and the total amount of Rab GTPase, we infer the saturation of Zfyve26 molecules at the endocytic surface from time-lapse imaging data. Our findings establish that transduction in the endocytic system is governed by stoichiometric constraints determining the trade-off between different trafficking pathways at the surface of a membrane-bound organelle.
Keywords: Cell biology; Functional aspects of cell biology; Molecular interaction; Organizational aspects of cell biology.
© 2025 The Author(s).
Conflict of interest statement
EH is a founder of Kither Biotech, a company involved in the development of PI3K inhibitors. E.H. and C.C.C. own patents related to quantification of Rab11 activity.
Figures




References
LinkOut - more resources
Full Text Sources

