Integrated bioinformatics analysis to develop diagnostic models for malignant transformation of chronic proliferative diseases
- PMID: 40201199
- PMCID: PMC11977743
- DOI: 10.1097/BS9.0000000000000226
Integrated bioinformatics analysis to develop diagnostic models for malignant transformation of chronic proliferative diseases
Abstract
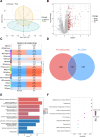
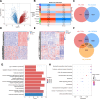
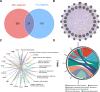
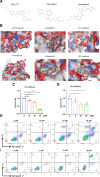
The combined analysis of dual diseases can provide new insights into pathogenic mechanisms, identify novel biomarkers, and develop targeted therapeutic strategies. Polycythemia vera (PV) is a chronic myeloproliferative neoplasm associated with a risk of acute myeloid leukemia (AML) transformation. However, the chronic nature of disease transformation complicates longitudinal high-throughput sequencing studies of patients with PV before and after AML transformation. This study aimed to develop a diagnostic model for malignant transformation of chronic proliferative diseases, addressing the challenges of early detection and intervention. Integrated public datasets of PV and AML were analyzed to identify differentially expressed genes (DEGs) and construct a weighted correlation network. Machine-learning algorithms screen genes for potential biomarkers, leading to the development of diagnostic models. Clinical specimens were collected to validate gene expression. cMAP and molecular docking predicted potential drugs. In vitro experiments were performed to assess drug efficacy in PV and AML cells. CIBERSORT and single-cell RNA-sequencing (scRNA-seq) analyses were used to explore the impact of hub genes on the tumor microenvironment. We identified 24 genes shared between PV and AML, which were enriched in immune-related pathways. Lactoferrin (LTF) and G protein-coupled receptor 65 (GPR65) were integrated into a nomogram with a robust predictive power. The predicted drug vemurafenib inhibited proliferation and increased apoptosis in PV and AML cells. TME analysis has linked these biomarkers to macrophages. Clinical samples were used to confirm LTF and GPR65 expression levels. We identified shared genes between PV and AML and developed a diagnostic nomogram that offers a novel avenue for the diagnosis and clinical management of AML-related PV.
Keywords: Acute myeloid leukemia; Bioinformatics analysis; Biomarker; Hub genes; Machine learning; Polycythemia vera.
Copyright © 2025 The Authors. Published by Wolters Kluwer Health Inc., on behalf of the Chinese Medical Association (CMA) and Institute of Hematology, Chinese Academy of Medical Sciences & Peking Union Medical College (IHCAMS).
Figures




References
LinkOut - more resources
Full Text Sources
Miscellaneous
