Survival path model outperforms conventional static machine learning models in long-term dynamic prognosis prediction for patients with intermediate stage hepatocellular carcinoma
- PMID: 40201235
- PMCID: PMC11978388
- DOI: 10.1093/bioadv/vbaf027
Survival path model outperforms conventional static machine learning models in long-term dynamic prognosis prediction for patients with intermediate stage hepatocellular carcinoma
Abstract
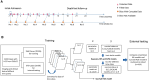
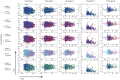
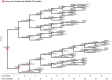
Motivation: Patients with intermediate stage hepatocellular carcinoma (HCC) require repeated disease monitoring, prognosis assessment, and treatment planning. A novel machine learning model called survival path mapping (SP) model was developed, while its performance as compared with conventional machine learning models remains unknown. Between January 2007 and December 2018, the time-series data of 2644 intermediate stage HCC patients from four medical centers in China were reviewed and included. Static machine learning models by Gaussian Naive Bayes (GNB), support vector machine (SVM), and random forest (RF) for the prediction of survivorship were built based on data at initial admission. Longitudinal data divided into different time slices were utilized for the construction of the SP model. The time-dependent c-index was compared between models.
Results: The training set, internal testing set, and external testing set consisted of 1560, 670, and 414 HCC patients, respectively. The survival path model had superior or non-inferior performance in prognosis prediction compared to GNB and RF models since the 12th month after initial diagnosis in the training set and the external testing set. The survival path model had higher time-dependent c-index over all conventional ML models since the 6th month in the external testing cohort. In conclusion, the survival path model had superior performance in long-term dynamic prognosis prediction compared to conventional static machine learning models for intermediate stage HCC.
Availability and implementation: The parameters of models are provided in the manuscript.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures

Similar articles
-
Machine Learning for Dynamic Prognostication of Patients With Hepatocellular Carcinoma Using Time-Series Data: Survival Path Versus Dynamic-DeepHit HCC Model.Cancer Inform. 2024 Oct 16;23:11769351241289719. doi: 10.1177/11769351241289719. eCollection 2024. Cancer Inform. 2024. PMID: 39421722 Free PMC article.
-
Development and validation of a machine learning-based predictive model for assessing the 90-day prognostic outcome of patients with spontaneous intracerebral hemorrhage.J Transl Med. 2024 Mar 4;22(1):236. doi: 10.1186/s12967-024-04896-3. J Transl Med. 2024. PMID: 38439097 Free PMC article.
-
Short-term prognosis for hepatocellular carcinoma patients with lung metastasis: A retrospective cohort study based on the SEER database.Medicine (Baltimore). 2022 Nov 11;101(45):e31399. doi: 10.1097/MD.0000000000031399. Medicine (Baltimore). 2022. PMID: 36397445 Free PMC article. Clinical Trial.
-
[Application value of machine learning algorithms for predicting recurrence after resection of early-stage hepatocellular carcinoma].Zhonghua Wai Ke Za Zhi. 2021 Aug 1;59(8):679-685. doi: 10.3760/cma.j.cn112139-20201026-00768. Online ahead of print. Zhonghua Wai Ke Za Zhi. 2021. PMID: 34192861 Chinese.
-
A machine learning-based prediction model pre-operatively for functional recovery after 1-year of hip fracture surgery in older people.Front Surg. 2023 Jun 7;10:1160085. doi: 10.3389/fsurg.2023.1160085. eCollection 2023. Front Surg. 2023. PMID: 37351328 Free PMC article.
References
-
- Brown ZJ, Tsilimigras DI, Ruff SM et al. Management of hepatocellular carcinoma: a review. JAMA Surg 2023;158:410–20. - PubMed
LinkOut - more resources
Full Text Sources

