Unraveling BRAF alterations: molecular insights to circumvent therapeutic resistance across cancer types
- PMID: 40201310
- PMCID: PMC11977354
- DOI: 10.20517/cdr.2024.213
Unraveling BRAF alterations: molecular insights to circumvent therapeutic resistance across cancer types
Abstract
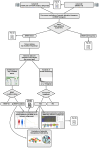
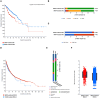
Aim: As intrinsic resistance - often driven by concurrent genomic alterations in tumor suppressor genes or oncogenes - remains a major challenge in oncology, this work aimed to comprehensively analyze BRAF somatic alterations across cancer types and identify new potential therapeutic strategies to overcome drug resistance. Methods: We conducted an extensive analysis of genomics, transcriptomics, and clinical data retrieved from public repositories, including cBioPortal. Our comprehensive analysis examined BRAF alterations [point mutations, structural variants (SVs) and copy number alteration] in more than 217,000 tumor samples across 120 distinct tumor types from primary and metastatic sites in both adult and pediatric cohorts, focusing on mutual exclusivity and co-occurrence of mutations in other oncogenes or tumor suppressors. The work also explores the association of BRAF somatic alterations with survival, clinical and molecular features. Results: Analysis of mutation frequencies across cancer types revealed that BRAFV600E represents approximately 90% of all BRAF alterations. While melanoma and thyroid carcinoma show the highest prevalence of BRAF mutations, followed by colorectal and non-small cell lung cancer in terms of absolute number of patients harboring BRAF mutations worldwide, notably high mutation frequencies were identified in rare malignancies, including hairy-cell leukemia, ganglioglioma, and serous borderline ovarian tumors. The comprehensive analysis of genomic profiling data across these tumors uncovered distinct patterns of co-occurring and mutually exclusive alterations in oncogenes and tumor suppressor genes, illuminating resistance mechanisms and suggesting novel therapeutic combinations. Conclusion: Comprehensive genomic profiling is critical for optimizing targeted therapy and overcoming drug resistance in BRAF-mutated cancers. The identification of co-occurring alterations provides opportunities for rational combination therapies, emphasizing the importance of detailed mutation profiling in developing effective treatment strategies across diverse cancer types.
Keywords: BRAF-mutated tumors; drug resistance; genomic profiling; mutation co-occurrence; mutually exclusive mutations; targeted therapy.
© The Author(s) 2025.
Conflict of interest statement
Giovannetti E is an Associate Editor on the Editorial Board of the journal Cancer Drug Resistance. Giovannetti E was not involved in any steps of editorial processing, notably including reviewer selection, manuscript handling, and decision making. The other authors declared that there are no conflicts of interest.
Figures



References
-
- Aprile M, Cataldi S, Perfetto C, Federico A, Ciccodicola A, Costa V. Targeting metabolism by B-raf inhibitors and diclofenac restrains the viability of BRAF-mutated thyroid carcinomas with Hif-1α-mediated glycolytic phenotype. Br J Cancer. 2023;129:249–65. doi: 10.1038/s41416-023-02282-2. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
