The epigenetic mechanisms involved in the treatment resistance of glioblastoma
- PMID: 40201311
- PMCID: PMC11977385
- DOI: 10.20517/cdr.2024.157
The epigenetic mechanisms involved in the treatment resistance of glioblastoma
Abstract
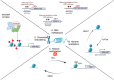
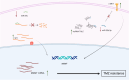
Glioblastoma (GBM) is an aggressive malignant brain tumor with almost inevitable recurrence despite multimodal management with surgical resection and radio-chemotherapy. While several genetic, proteomic, cellular, and anatomic factors interplay to drive recurrence and promote treatment resistance, the epigenetic component remains among the most versatile and heterogeneous of these factors. Herein, the epigenetic landscape of GBM refers to a myriad of modifications and processes that can alter gene expression without altering the genetic code of cancer cells. These processes encompass DNA methylation, histone modification, chromatin remodeling, and non-coding RNA molecules, all of which have been found to be implicated in augmenting the tumor's aggressive behavior and driving its resistance to therapeutics. This review aims to delve into the underlying interactions that mediate this role for each of these epigenetic components. Further, it discusses the two-way relationship between epigenetic modifications and tumor heterogeneity and plasticity, which are crucial to effectively treat GBM. Finally, we build on the previous characterization of epigenetic modifications and interactions to explore specific targets that have been investigated for the development of promising therapeutic agents.
Keywords: DNA methylation; epigenetics; glioblastoma; histone modification; miRNA; treatment resistance; tumoral heterogeneity.
© The Author(s) 2025.
Conflict of interest statement
Tyler B has research funding from NIH. Therapeutics A Inc. has licensed one of her patents, and she holds stock in Peabody Pharmaceuticals* (*including equity or options). The other authors declared that there are no conflicts of interest.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
