Enhanced analysis of the genomic diversity of Mycobacterium bovis in Great Britain to aid control of bovine tuberculosis
- PMID: 40201440
- PMCID: PMC11975571
- DOI: 10.3389/fmicb.2025.1515906
Enhanced analysis of the genomic diversity of Mycobacterium bovis in Great Britain to aid control of bovine tuberculosis
Abstract
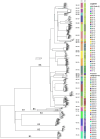
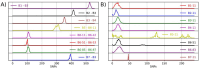
Bovine tuberculosis (bTB) is an endemic disease in Great Britain (GB) that affects mainly cattle but also other livestock and wild mammal species, leading to significant economic and social impact. Traditional genotyping of Mycobacterium bovis (M. bovis) isolates, which cause bTB, had been used routinely since the late 1990s as the main resource of genetic information in GB to describe their population and to understand their epidemiology. Since 2017, whole-genome sequencing (WGS) has been implemented on M. bovis isolates collected during routine surveillance. In this study, we analysed genome sequences from 3,052 M. bovis isolates from across GB to characterise their diversity and population structure in more detail. Our findings show that the M. bovis population in GB, based on WGS, is more diverse than previously indicated by traditional genotyping and can be divided into seven major clades, with one of them subdivided further into 29 clades that differ from each other by at least 70 single-nucleotide polymorphisms (SNPs). Based on the observed phylogenetic structure, we present a SNP-based classification system that replaces the genotype scheme that had been used until recently in GB. The predicted function and associated processes of the genes harbouring these SNPs are discussed with potential implications for phenotypic/functional differences between the identified clades. At the local scale, we show that WGS provides greater discriminatory power and that it can reveal the origin of infection and associated risk pathways even in areas of high bTB prevalence. The difficulty in determining transmission pathways due to the limited discrimination of isolates by traditional typing methods has compromised bTB control, as without such information it is harder to determine the relative efficacy of potential intervention measures. This study demonstrates that the higher resolution provided by WGS data can improve determination of infection sources and transmission pathways, provide important insights that will inform and shape bTB control policies in GB, as well as improve farm specific advice on interventions that are likely to be effective.
Keywords: Mycobacterium bovis; bovine tuberculosis (bTB); clade; epidemiology; phylogenetics; whole genome sequencing.
Copyright © 2025 Sandhu, Nunez-Garcia, Berg, Wheeler, Dale, Upton, Gibbens, Hewinson, Downs, Ellis and Palkopoulou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Aínsa J. A., Ryding N. J., Hartley N., Findlay K. C., Bruton C. J., Chater K. F. (2000). WhiA, a protein of unknown function conserved among gram-positive bacteria, is essential for sporulation in Streptomyces coelicolor A3(2). J. Bacteriol. 182, 5470–5478. doi: 10.1128/jb.182.19.5470-5478.2000, PMID: - DOI - PMC - PubMed
-
- Allen A. R., Skuce R. A., Byrne A. W. (2018). Bovine tuberculosis in Britain and Ireland – A perfect storm? The confluence of potential ecological and epidemiological impediments to controlling a chronic infectious disease. Front. Vet. Sci. 5:109. doi: 10.3389/fvets.2018.00109, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

