Ethylene-Activated E3 Ubiquitin Ligase MdEAEL1 Promotes Apple Fruit Softening by Facilitating the Dissociation of Transcriptional Repressor Complexes
- PMID: 40202115
- PMCID: PMC12165066
- DOI: 10.1002/advs.202417393
Ethylene-Activated E3 Ubiquitin Ligase MdEAEL1 Promotes Apple Fruit Softening by Facilitating the Dissociation of Transcriptional Repressor Complexes
Abstract
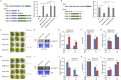
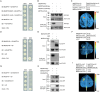
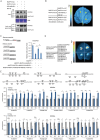
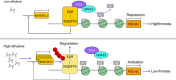
Fruit of most apple varieties soften after harvest, and although the hormone ethylene is known to induce softening, the associated pathway is not well resolved. In this study, it is determined that MdEAEL1 (Ethylene-activated E3 ubiquitin Like 1) is specifically expressed during apple fruit postharvest storage, activated by ethylene, and interacts with the transcription factor MdZFP3 (zinc finger protein3). MdZFP3 is found to rely on an EAR (ethylene-responsive element binding factor-associated amphiphilic repression) motif to form a transcriptional repression complex with MdTPL4 (TOPLESS4)-MdHDA19 (histone deacetylase19), thereby downregulating the histone acetylation levels of the promoters of a range of cell wall degradation-related genes and inhibiting their transcription. MdEAEL1 ubiquitinates and degrades MdZFP3, leading to the disassembly of the MdZFP3-MdTPL4-MdHDA19 transcriptional repression complex. This process promotes the transcription of cell wall degradation-related genes, resulting in fruit softening during storage. Furthermore, the disassembly of the MdZFP3-MdTPL4-MdHDA19 transcriptional repression complex, mediated by MdEAEL1, upregulates the transcription of MdEAEL1 itself, creating a feedback loop that further promotes softening. This study elucidates the interplay between post-translational modifications of a transcription factor and its epigenetic modification to regulate fruit softening, and highlights the complexity of ethylene-induced softening.
Keywords: apple fruit; ethylene; fruit softening.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Marín‐Rodríguez M. C., Orchard J., Seymour G. B., J. Exp. Bot. 2002, 53, 2115. - PubMed
-
- Wang D., Yeats T. H., Uluisik S., Rose J. K. C., Seymour G. B., Trends Plant Sci. 2018, 23, 302. - PubMed
-
- a) Tucker G., Yin X., Zhang A., Wang M., Zhu Q., Liu X., Xie X., Chen K., Grierson D., Food Qual. Saf. 2017, 1, 253;
- b) Rose J. K. C., Bennett A. B., Trends Plant Sci. 1999, 4, 176. - PubMed
-
- a) King G. A., O'Donoghue E. M., Trends Food Sci. Technol. 1995, 6, 385;
- b) Brummell D. A., Bowen J. K., Gapper N. E., Curr. Opin. Biotechnol. 2022, 78, 102786. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
