A trans-Translation Inhibitor is Potentiated by Zinc and Kills Mycobacterium tuberculosis and Nontuberculous Mycobacteria
- PMID: 40202906
- PMCID: PMC12071686
- DOI: 10.1021/acsinfecdis.4c00963
A trans-Translation Inhibitor is Potentiated by Zinc and Kills Mycobacterium tuberculosis and Nontuberculous Mycobacteria
Abstract
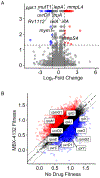
Mycobacterium tuberculosis poses a serious challenge for human health, and new antibiotics with novel targets are needed. Here we demonstrate that an acylaminooxadiazole, MBX-4132, specifically inhibits the trans-translation ribosome rescue pathway to kill M. tuberculosis. Our data demonstrate that MBX-4132 is bactericidal against multiple pathogenic mycobacterial species and kills M. tuberculosis in macrophages. We also show that acylaminooxadiazole activity is antagonized by iron but is potentiated by zinc. Our transcriptomic data reveal dysregulation of multiple metal homeostasis pathways after exposure to MBX-4132. Furthermore, we see differential expression of genes related to zinc sensing and efflux when trans-translation is inhibited. Taken together, these data suggest that there is a link between disturbing intracellular metal levels and acylaminooxadiazole-mediated inhibition of trans-translation. These findings provide an important proof-of-concept that trans-translation is a promising antitubercular drug target.
Keywords: Mycobacterium tuberculosis; antibiotic resistance; metal homeostasis; nontuberculous mycobacteria; oxadiazole; trans-translation.
Conflict of interest statement
DECLARATIONS OF INTERESTS
Kenneth Keiler is an inventor on the patent for MBX-4132.
Figures




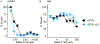

Update of
-
A trans-translation inhibitor is potentiated by zinc and kills Mycobacterium tuberculosis and non-tuberculous mycobacteria.bioRxiv [Preprint]. 2024 Nov 2:2024.11.02.621434. doi: 10.1101/2024.11.02.621434. bioRxiv. 2024. Update in: ACS Infect Dis. 2025 May 9;11(5):1140-1152. doi: 10.1021/acsinfecdis.4c00963. PMID: 39554143 Free PMC article. Updated. Preprint.
References
-
- World Health Organization, WHO Global Tuberculosis Program. Global Tuberculosis Report 2024. World Health Organization, 2024. https://iris.who.int/bitstream/handle/10665/379339/9789240101531-eng.pdf... (accessed 2025-03-25)
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

