Rapid bacterial identification and resistance detection using a low complexity molecular diagnostic platform in Zimbabwe
- PMID: 40202992
- PMCID: PMC11981161
- DOI: 10.1371/journal.pgph.0004343
Rapid bacterial identification and resistance detection using a low complexity molecular diagnostic platform in Zimbabwe
Abstract
Background: Sepsis is a major cause of mortality in low-resource settings. Effective microbiological culture services are a bottleneck in diagnosis and surveillance.
Aim: We aimed to evaluate the performance of the BIOFIRE FILMARRAY Blood Culture Identification 2 (BCID2, bioMérieux) assay in a low-resource setting laboratory in comparison to standard practice.
Methods: This five month prospective validation study included all positive blood cultures collected at Sally Mugabe Central Hospital, Harare, Zimbabwe. BCID2 testing was done in parallel to standard phenotypic procedures and resistance testing. Reference identification was performed using mass spectrometry or whole genome sequencing. Only samples with available reference standard results were included in the analysis. Data captured on paper-based forms was entered into electronic case report forms (ODK Collect). Specificity and sensitivity for BCID2 were calculated in comparison to the reference standards, with performance measures calculated using the Wilson score. Biomedical scientists using BCID2 completed a system usability survey (SUS).
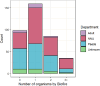
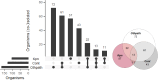
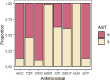
Results: Positive results were recorded in 780/2,023 (38.5%) blood cultures, within which 377 (48.3%) had reference results and so were included in analysis. Neonatal samples were most frequent (182, 48.3%), then paediatric (150, 39.8%), then adults (18, 4.8%) and unknown (27, 7.2%). Specificity exceeded 95% throughout. Sensitivity ranged from 50% (A. calcoaceticus-baumanii complex, Proteus spp.) to 100% (S. pneumoniae, Salmonella spp). Using BCID2, CTX-M was detected in 111/175 (74.5%) Enterobacterales, from which 5/111 also had NDM and VIM detected. NDM-5 was detected in 2/5 NDM samples using sequencing. In total 3/23 S. aureus isolates were methicillin resistant, from which one was confirmed using phenotypic antimicrobial susceptibility testing. Usability was good (SUS score = 79.5).
Conclusion: Rapid molecular tests have potential to improve turn-around time and quality of sepsis diagnostics. However, specific work-flows are critical to supplement molecular tests with minimal phenotypic tests for optimal clinical decision-making.
Copyright: © 2025 Mwaturura et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
LinkOut - more resources
Full Text Sources
Miscellaneous
