Distinctive Membrane Accommodation Traits Underpinning the Neutralization Activity of HIV-1 Antibody against MPER
- PMID: 40202993
- PMCID: PMC12056697
- DOI: 10.1021/acs.molpharmaceut.4c01341
Distinctive Membrane Accommodation Traits Underpinning the Neutralization Activity of HIV-1 Antibody against MPER
Abstract
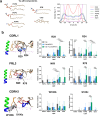
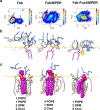
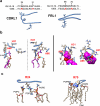
The membrane-proximal external region (MPER), located in the carboxy-terminal section of HIV's envelope glycoprotein (Env) ectodomain, which is essential for viral entry into host cells, has gained considerable attention as a target for HIV vaccine development due to the exceptional neutralization breadth of antibodies against MPER epitopes. A distinctive feature of broadly neutralizing antibodies (bnAbs) targeting MPER is their requirement to accommodate the viral membrane into the surface of the antigen-binding fragment, or Fab moiety, to optimize antigen recognition. In this study, we sought to elucidate the molecular mechanism behind this interaction and its relevance to the antiviral function of bnAb 10E8. We conducted all-atom molecular dynamics simulations of three systems: (i) Fab 10E8 positioned on the surface of a viral-like lipid bilayer (VL-LB), (ii) Fab 10E8 in complex with an MPER helix anchored to the VL-LB via the Env glycoprotein transmembrane domain (TMD), and (iii) a Fab/MPER-TMD complex similarly embedded in the VL-LB but with a chemically optimized Fab 10E8 variant showing enhanced potency. Comparing these systems enabled us to derive atomic-scale Fab-membrane accommodation profiles pertinent to 10E8's neutralizing function. Our findings support that Fab adaptation to the viral membrane interface following epitope binding is crucial for developing MPER-targeted neutralizing activity. This analysis also provides insights into pathways for strengthening lipid interactions, which may prove valuable in designing MPER-based biologics and vaccines to prevent or treat HIV infection.
Keywords: HIV-1 neutralization mechanism; MD simulations; MPER antibody; antibody engineering; antibody–membrane interaction; site-selective chemical modification; therapeutic antibody.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
-
- Kunert R.; Wolbank S.; Stiegler G.; Weik R.; Katinger H. Characterization of molecular features, antigen-binding, and in vitro properties of IgG and IgM variants of 4E10, an anti-HIV type 1 neutralizing monoclonal antibody. AIDS Res. Hum. Retroviruses 2004, 20 (7), 755–762. 10.1089/0889222041524571. - DOI - PubMed
-
- Pinto D.; Fenwick C.; Caillat C.; Silacci C.; Guseva S.; Dehez F.; Chipot C.; Barbieri S.; Minola A.; Jarrossay D.; et al. Structural basis for broad HIV-1 neutralization by the MPER-specific human broadly neutralizing antibody LN01. Cell Host Microbe 2019, 26 (5), 623–637. 10.1016/j.chom.2019.09.016. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

