Genome-wide analysis of fitness determinants of Staphylococcus aureus during growth in milk
- PMID: 40203072
- PMCID: PMC12011298
- DOI: 10.1371/journal.ppat.1013080
Genome-wide analysis of fitness determinants of Staphylococcus aureus during growth in milk
Abstract
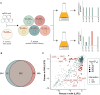
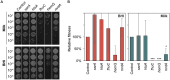
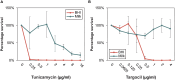
Staphylococcus aureus is a major concern in the dairy industry due to its significance as a pathogen causing bovine mastitis as well as a source of food poisoning. The nutrient-rich milk environment supports bacterial growth, but the specific genetic determinants that facilitate S. aureus proliferation and persistence in milk are poorly understood. In this study, we conducted a genome-wide CRISPR interference sequencing (CRISPRi-seq) screen with the laboratory strain S. aureus NCTC8325-4, to identify fitness determinants essential for S. aureus growth and survival in milk. We identified 282 milk-essential genes, including those with key roles in DNA replication, protein synthesis, and metabolism. Comparative analysis with brain heart infusion (BHI) as growth medium, revealed 79 genes with differential fitness, highlighting specific adaptations required for growth in milk. Notably, we found that purine biosynthesis, folate cycle pathways, and metal acquisition were particularly important in this environment. Based on this, we further demonstrate that S. aureus is more sensitive to the folate inhibitors trimethoprim-sulfamethoxazole (TMP-SMX) in milk and identify several genes whose knockdown results in hypersensitivity to TMP-SMX in milk. Additionally, our analysis showed a relatively reduced importance of cell wall components, such as teichoic acids, for S. aureus fitness in milk, which is also reflected in reduced efficiency of antimicrobials targeting teichoic acids. Together, these findings provide new insights into the genetic basis of S. aureus fitness and antibiotic susceptibility in milk, offering directions for novel treatment strategies against bovine mastitis.
Copyright: © 2025 Mårli et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Riva A, Borghi E, Cirasola D, Colmegna S, Borgo F, Amato E, et al. Methicillin-resistant Staphylococcus aureus in raw milk: prevalence, SCCmec typing, enterotoxin characterization, and antimicrobial resistance patterns. J Food Prot. 2015;78(6):1142–6. doi: 10.4315/0362-028X.JFP-14-531 - DOI - PubMed
-
- Marshall VM, Tamime AY. Starter cultures employed in the manufacture of biofermented milks. Int J of Dairy Tech. 1997;50(1):35–41. doi: 10.1111/j.1471-0307.1997.tb01733.x - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

