ARMC1 partitions between distinct complexes and assembles MIRO with MTFR to control mitochondrial distribution
- PMID: 40203102
- PMCID: PMC11980836
- DOI: 10.1126/sciadv.adu5091
ARMC1 partitions between distinct complexes and assembles MIRO with MTFR to control mitochondrial distribution
Abstract
Maintaining an optimal mitochondrial distribution is critical to ensure an adequate supply of energy and metabolites to support important cellular functions. How cells balance dynamic mitochondrial processes to achieve homeostasis is incompletely understood. Here, we show that ARMC1 partitioning between distinct mitochondrial protein complexes is a key determinant of mitochondrial distribution. In one complex, the mitochondrial trafficking adaptor MIRO recruits ARMC1, which mediates the assembly of a mitochondrial fission regulator (MTFR). MTFR stability depends on ARMC1, and MIRO-MTFR complexes specifically antagonize retrograde mitochondrial movement. In another complex, DNAJC11 facilitates ARMC1 release from mitochondria. Disrupting MIRO-MTFR assembly fails to rescue aberrant mitochondrial distributions clustered in the perinuclear area observed with ARMC1 deletion, while disrupting ARMC1 interaction with DNAJC11 leads to excessive mitochondrially localized ARMC1 and distinct mitochondrial defects. Thus, the abundance and trafficking impact of MIRO-MTFR complexes require ARMC1, whose mito-cytoplasmic shuttling balanced by DNAJC11 tunes steady-state mitochondrial distributions.
Figures
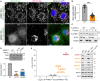


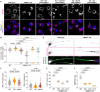
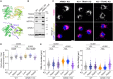

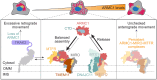
References
-
- Fransson Å., Ruusala A., Aspenström P., The atypical Rho GTPases Miro-1 and Miro-2 have essential roles in mitochondrial trafficking. Biochem. Biophys. Res. Commun. 344, 500–510 (2006). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

