Computer vision-guided rapid and precise automated cranial microsurgeries in mice
- PMID: 40203110
- PMCID: PMC11980847
- DOI: 10.1126/sciadv.adt9693
Computer vision-guided rapid and precise automated cranial microsurgeries in mice
Abstract
A common procedure that allows interfacing with the brain is cranial microsurgery, wherein small to large craniotomies are performed on the overlying skull for insertion of neural interfaces or implantation of optically clear windows for long-term cranial observation. Performing craniotomies requires skill, time, and precision to avoid damaging the brain and dura. Here, we present a computer vision-guided craniotomy robot (CV-Craniobot) that uses machine learning to accurately estimate the dorsal skull anatomy from optical coherence tomography images. Instantaneous information of skull morphology is used by a robotic mill to rapidly and precisely remove the skull from a desired craniotomy location. We show that the CV-Craniobot can perform small (2- to 4-millimeter diameter) craniotomies with near 100% success rates within 2 minutes and large craniotomies encompassing most of the dorsal cortex in less than 10 minutes. Thus, the CV-Craniobot enables rapid and precise craniotomies, reducing surgery time compared to human practitioners and eliminating the need for long training.
Figures


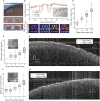
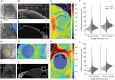
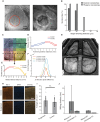
References
-
- Yu C.-H., Yu Y., Adsit L. M., Chang J. T., Barchini J., Moberly A. H., Benisty H., Kim J., Young B. K., Heng K., Farinella D. M., Leikvoll A., Pavan R., Vistein R., Nanfito B. R., Hildebrand D. G. C., Otero-Coronel S., Vaziri A., Goldberg J. L., Ricci A. J., Fitzpatrick D., Cardin J. A., Higley M. J., Smith G. B., Kara P., Nielsen K. J., Smith I. T., Smith S. L., The Cousa objective: A long-working distance air objective for multiphoton imaging in vivo. Nat. Methods 21, 132–141 (2024). - PMC - PubMed
-
- Hope J., Beckerle T. M., Cheng P.-H., Viavattine Z., Feldkamp M., Fausner S. M. L., Saxena K., Ko E., Hryb I., Carter R. E., Ebner T. J., Kodandaramaiah S. B., Brain-wide neural recordings in mice navigating physical spaces enabled by robotic neural recording headstages. Nat. Methods 21, 2171–2181 (2024). - PubMed
-
- Bennett C., Ouellette B., Ramirez T. K., Cahoon A., Cabasco H., Browning Y., Lakunina A., Lynch G. F., McBride E. G., Belski H., Gillis R., Grasso C., Howard R., Johnson T., Loeffler H., Smith H., Sullivan D., Williford A., Caldejon S., Durand S., Gale S., Guthrie A., Ha V., Han W., Hardcastle B., Mochizuki C., Sridhar A., Suarez L., Swapp J., Wilkes J., Siegle J. H., Farrell C., Groblewski P. A., Olsen S. R., SHIELD: Skull-shaped hemispheric implants enabling large-scale electrophysiology datasets in the mouse brain. Neuron 112, 2869–2885.e8 (2024). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

