Quantitative Proteomics and Phosphoproteomics Analysis of Patient-Derived Ovarian Cancer Stem Cells
- PMID: 40204276
- PMCID: PMC12142526
- DOI: 10.1016/j.mcpro.2025.100965
Quantitative Proteomics and Phosphoproteomics Analysis of Patient-Derived Ovarian Cancer Stem Cells
Abstract
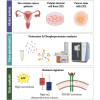
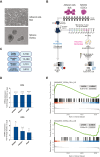
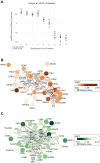
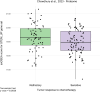
High-grade serous ovarian carcinoma (HGSOC) is the deadliest gynecologic cancer. Key to the progression and ultimate lethality of this subtype is the intra-tumoral heterogeneity, which is defined as the coexistence of different cell types and populations within a single tumor. Among those, ovarian cancer stem cells (OCSCs) are a distinct subpopulation of tumor cells endowed with stem-like properties, which can survive current standard therapies, resulting in tumor recurrence. Here, we generated ex vivo primary OCSC-enriched three-dimensional (3D) spheres from 10 distinct treatment naive patient-derived adherent (2D) cultures. We used state-of-the-art quantitative mass spectrometry to characterize the molecular events associated with OCSCs by analyzing their proteome and phosphoproteome. Our data revealed a stemness-related protein signature, shared within a heterogeneous patient cohort, which correlates with chemo-refractoriness in a clinical proteomics dataset. Moreover, we identified targetable deregulated kinases and aberrant PDGF receptor activation in OCSCs. Pharmacological inhibition of PDGFR in adherent OC cells reduced the stemness potential, measured by sphere formation assay. Overall, we provide a valuable resource to identify new OCSC markers and putative targets for OCSC-directed therapies.
Keywords: HGSOC; cancer stem cells; ovarian cancer; phosphoproteomics; proteomics.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures


References
-
- Martinez-Val A., Guzmán U.H., Olsen J.V. Obtaining complete human proteomes. Annu. Rev. Genomics Hum. Genet. 2022 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

